













| General Information: |
|
| Name(s) found: |
GGA2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 585 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSHPHSHSIY LSELPVRKPQ ALGNPLLRKI QRACRMSLAE PDLALNLDIA DYINEKQGAA 60
61 PRDAAIALAK LINNRESHVA IFALSLLDVL VKNCGYPFHL QISRKEFLNE LVKRFPGHPP 120
121 LRYSKIQRLI LTAIEEWYQT ICKHSSYKND MGYIRDMHRL LKYKGYAFPK ISESDLAVLK 180
181 PSNQLKTASE IQKEQEIAQA AKLEELIRRG KPEDLREANK LMKIMAGFKE DNAVQAKQAI 240
241 SSELNKLKRK ADLLNEMLES PDSQNWDNET TQELHSALKV AQPKFQKIIE EEQEDDALVQ 300
301 DLLKFNDTVN QLLEKFNLLK NGDSNAASQI HPSHVSAPLQ QSSGALTNEI NLIDFNDLDE 360
361 APSQGNNNTN GTGTPAAAET SVNDLLGDLT DLSISNPSTA NQASFGLGGD IVLGSSQPAP 420
421 PVTTTNNSNN TLDLLGLSTP QSPTNSQAVN SSGFDLLMGF NPTTGTTTAP ARTLVNQSPN 480
481 LKIEFEISRE SNSVIRIKSF FTNLSSSPIS NLVFLLAVPK SMSLKLQPQS SNFMIGNAKD 540
541 GISQEGTIEN APANPSKALK VKWKVNYSVN STQAEETAVF TLPNV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
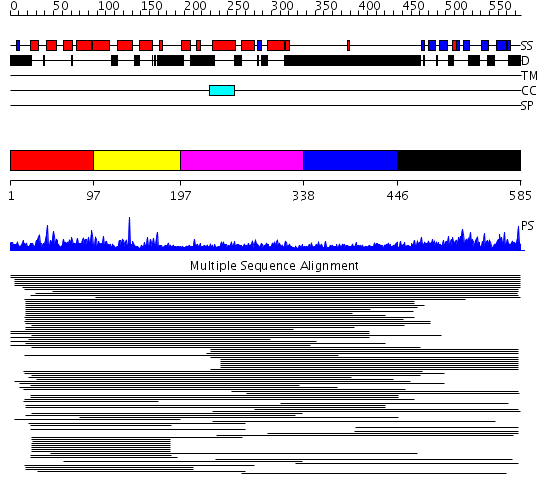
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | 177.626727 | Tom1 protein | |
| 2 | View Details | [97..196] | 177.626727 | Tom1 protein | |
| 3 | View Details | [197..337] | 3.0 | GAT domain No confident structure predictions are available. | |
| 4 | View Details | [338..445] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [446..585] | 36.522879 | Gamma1-adaptin domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)