













| General Information: |
|
| Name(s) found: |
FYV8_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 817 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTEQVGRKK SYRWVSASQA SYDGAGWDSS DEYDYSSEDG TKGSEIHKQK ISNLPSLPKL 60
61 NYTDVNGEHD ENTGENKDSN DNNVSKSDIS PSDKEVGYLS DGVPKLMASR ESVELQAKKS 120
121 SEHSKSDYLS STASLKSPSE NKKSPHTNRA VNEDLDNLIE QISREMTPEI RQTSDFRRDS 180
181 DSCDEIQNEA PLGEAVPSSS SPVEEDEKSH SLGVSMDTNE ADTTFNTPTR NGNEHLSSDG 240
241 DVSEQKDDEF KVSERGYLAD ILPAEKEENL QQEDDGEVES SGALEKKEKS EEKTSIRNRN 300
301 STSSGQDKVA KPKPVANETK TSDNGYRNSF FNDYQHSSDS EEDDNNEGNS GSSDDDNRSS 360
361 VSDKHADINR QSKQLDTTDD DALSYTESIK YSTNETEEED NEDNESIEDK NEDNESIEDE 420
421 NEDTDSYKFS NREKGSILLT SDEEEEEKGM SSDSDEGSLK APKSGYFSKM IGNDDKGDSA 480
481 LQPNQIDTIE NTNLSNSGSE LENSDGSDEE DHINEDKVLE ESSVKDSTDV DSWKPDSEAL 540
541 RSGFVQDTAN KKAPPGYVID SNGKLVDLTP ASMKPRVVST YSEMESTWDA FPSKGEDDDL 600
601 ETIRDTKTIY DNNTIYNVPG LIGNQSNLPP LPMDAQEQLN AGNDNSTTDN DNSNNTANDL 660
661 AARSASFKSE NRTVSQGEMT SVHEPSTEEM AKLGQQNNLP KLDMNKLLNS KTSHAGKIEQ 720
721 LRNYKRELDE YDTGIQTWIN YTLKSSSNKD KDFIAEEYKQ HSHVREAYAN ADDLSKKHTV 780
781 INTVASVNQN VTHLRRKVFQ HSMKPKDLFA SIGKKKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
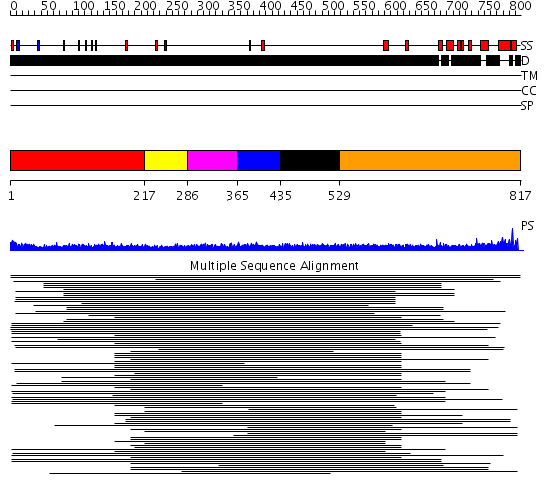
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..216] | 7.38 | No description for 1ldvA was found. | |
| 2 | View Details | [217..285] | 12.52 | No description for 1l8mA was found. | |
| 3 | View Details | [286..364] | 12.52 | No description for 1l8mA was found. | |
| 4 | View Details | [365..434] | 12.52 | No description for 1l8mA was found. | |
| 5 | View Details | [435..528] | 12.52 | No description for 1l8mA was found. | |
| 6 | View Details | [529..817] | 2.192997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)