













| General Information: |
|
| Name(s) found: |
FUS2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 677 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFKTSYNLYD LNYPKNDSLT PIRDYKNDYF HKNDDKLPEI VRKPTRKLSK HENKLNDKKF 60
61 TNKRPASLDL HSIVESLSNK KIYSPINTEI FQNVVRLNLS PQIPNSPHEG CKFYKIVQEF 120
121 YLSEVEYYNN LLTANNVYRK ALNSDPRFKN KLVKLDSSDE LLLFGNIDTI ASISKILVTA 180
181 IKDLLLAKQR GKMLDANEWQ KIFTKNEVQQ QLYSTFDISE AFEQHLLRIK STYTSYFVSH 240
241 QKQMELFTTL RMNKNHFFNK WYEYCLKESG CIKLEDILKS PMKRLTQWID TLETLESCYE 300
301 DILSPELGLK LSPTRRKYSL FSNKLETEVS EYKSNSMYNF SLTPSEIIQS YDEDQFTHLL 360
361 KPPDKQNKNI CNASRQESNL DNSRVPSLLS GSSSYYSDVS GLEIVTNTST ASAEMINLKM 420
421 DEETEFFTLA DHISKFKKVM KGLLELKKNL LKNDLSGIID ISLRRINAWK KVIECERPSG 480
481 AFFAHDNLIS TMCSSYIDKL HEQKNQVTIL KLTELETDVM NPLERIIAHC TTVKSKLKDL 540
541 QALKKDYMLF LQEKKANVRD IKRDLLGMHF QNLQNQMKRE LPVFITLIHD TIECILLNYI 600
601 KVFLKYLEII AGGKKYLQKD LENMSLNDSI ATGQIKNLDI LQCYSKSRYM TKRMVRKDWP 660
661 FPGDPSGSRV VRKLFEL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
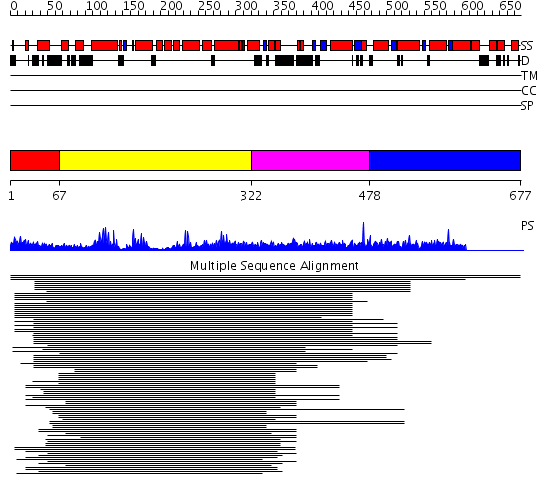
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..66] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [67..321] | 40.69897 | Dbl's big sister, Dbs | |
| 3 | View Details | [322..477] | 40.69897 | Dbl's big sister, Dbs | |
| 4 | View Details | [478..677] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 | No functions predicted. | ||||||||||||
| 3 |
|
||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)