













| General Information: |
|
| Name(s) found: |
DUG2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 878 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYDSRGVALH SELIHRWNHA FSILSIVAFP KKRLLFAGSQ DSKILVFDLP TYNLIHTIRL 60
61 GESQEETHTR SSVLCLTGSE DENFLFSGGA DSLVRIWSIG EKTIRDDFLP VTEIATVYSV 120
121 TDIGDIFSLA YLDSLETIVF GCQNASLLYV ENLIQKIEKK SSDGVENINK LPHRRYDKFF 180
181 DSLGPTGYSS NSLSQTSLTS LQENCGAAII EVPSENIIKY AHYGFIYSIN KLCPRFNQLL 240
241 EKSSRTSGAE HIISSAGDGI SKLWEFSKDK GQNTVKISLI NDKIDNEDSV ISQTIEFPFL 300
301 YCGLTDGIIK IWDLNTQQII STLKTKHESD VISISVYMDH VFAIDESGIT HFYQNQVNHW 360
361 NPQQGKILSS EIFSKSNAGS VSLLTGGSDG SLTLWDITSL LSAVPLSSNS PINASSTLQT 420
421 TNLWAAYQSA SLNNEEMLNT LRELISFQTV SQSKDTTNTL SLRRCAIYLQ QLFLKFGATN 480
481 SQLFPLPDGG NPVVFAYFQG NGKVSQVKGA KKKRILWYGH YDVISSGNTF NWNTDPFTLT 540
541 CENGYLKGRG VSDNKGPLVS AIHSVAYLFQ QGELVNDVVF LVEGSEEIGS ASLKQVCEKY 600
601 HDIIGKDIDW ILLSNSTWVD QEHPCLNYGL RGVINAQIKV WSDKPDGHSG LNGGVYDEPM 660
661 VNLVKIVSKL QNEQNEIMIP NFYSPLKDLT EEEYQRFQKI TELANIDENT TVQDLITNWT 720
721 KPSLSMTTVK FSGPGNITVI PKSVTMGISI RLVPEQSVEQ VKRDLKAYLE ESFKQLKSQN 780
781 HLEIKVLNEA EGWLGDPTNH AYQILKDEIT TAWDVEPLLV REGGSISCLR MLERIFDAPA 840
841 VQIPCGQSTD NGHLANENLR IKNWSNLTEI LSKVFNRL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
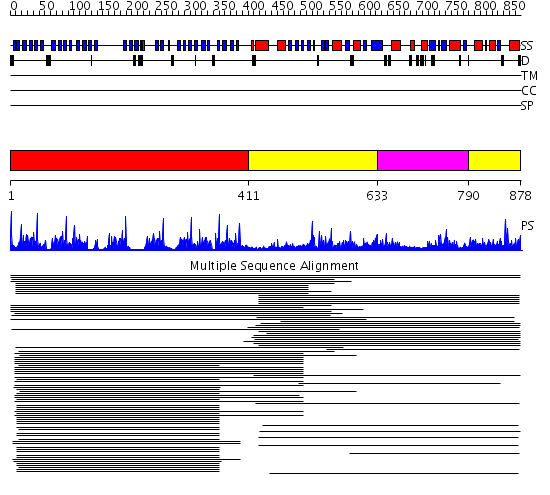
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..410] | 81.221849 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 2 | View Details | [411..632] [790..878] |
28.69897 | Carboxypeptidase G2, catalytic domain; Carboxypeptidase G2 | |
| 3 | View Details | [633..789] | 28.69897 | Carboxypeptidase G2, catalytic domain; Carboxypeptidase G2 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)