













| General Information: |
|
| Name(s) found: |
DNA2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1522 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPGTPQKNKR SASISVSPAK KTEEKEIIQN DSKAILSKQT KRKKKYAFAP INNLNGKNTK 60
61 VSNASVLKSI AVSQVRNTSR TKDINKAVSK SVKQLPNSQV KPKREMSNLS RHHDFTQDED 120
121 GPMEEVIWKY SPLQRDMSDK TTSAAEYSDD YEDVQNPSST PIVPNRLKTV LSFTNIQVPN 180
181 ADVNQLIQEN GNEQVRPKPA EISTRESLRN IDDILDDIEG DLTIKPTITK FSDLPSSPIK 240
241 APNVEKKAEV NAEEVDKMDS TGDSNDGDDS LIDILTQKYV EKRKSESQIT IQGNTNQKSG 300
301 AQESCGKNDN TKSRGEIEDH ENVDNQAKTG NAFYENEEDS NCQRIKKNEK IEYNSSDEFS 360
361 DDSLIELLNE TQTQVEPNTI EQDLDKVEKM VSDDLRIATD STLSAYALRA KSGAPRDGVV 420
421 RLVIVSLRSV ELPKIGTQKI LECIDGKGEQ SSVVVRHPWV YLEFEVGDVI HIIEGKNIEN 480
481 KRLLSDDKNP KTQLANDNLL VLNPDVLFSA TSVGSSVGCL RRSILQMQFQ DPRGEPSLVM 540
541 TLGNIVHELL QDSIKYKLSH NKISMEIIIQ KLDSLLETYS FSIIICNEEI QYVKELVMKE 600
601 HAENILYFVN KFVSKSNYGC YTSISGTRRT QPISISNVID IEENIWSPIY GLKGFLDATV 660
661 EANVENNKKH IVPLEVKTGK SRSVSYEVQG LIYTLLLNDR YEIPIEFFLL YFTRDKNMTK 720
721 FPSVLHSIKH ILMSRNRMSM NFKHQLQEVF GQAQSRFELP PLLRDSSCDS CFIKESCMVL 780
781 NKLLEDGTPE ESGLVEGEFE ILTNHLSQNL ANYKEFFTKY NDLITKEESS ITCVNKELFL 840
841 LDGSTRESRS GRCLSGLVVS EVVEHEKTEG AYIYCFSRRR NDNNSQSMLS SQIAANDFVI 900
901 ISDEEGHFCL CQGRVQFINP AKIGISVKRK LLNNRLLDKE KGVTTIQSVV ESELEQSSLI 960
961 ATQNLVTYRI DKNDIQQSLS LARFNLLSLF LPAVSPGVDI VDERSKLCRK TKRSDGGNEI1020
1021 LRSLLVDNRA PKFRDANDDP VIPYKLSKDT TLNLNQKEAI DKVMRAEDYA LILGMPGTGK1080
1081 TTVIAEIIKI LVSEGKRVLL TSYTHSAVDN ILIKLRNTNI SIMRLGMKHK VHPDTQKYVP1140
1141 NYASVKSYND YLSKINSTSV VATTCLGIND ILFTLNEKDF DYVILDEASQ ISMPVALGPL1200
1201 RYGNRFIMVG DHYQLPPLVK NDAARLGGLE ESLFKTFCEK HPESVAELTL QYRMCGDIVT1260
1261 LSNFLIYDNK LKCGNNEVFA QSLELPMPEA LSRYRNESAN SKQWLEDILE PTRKVVFLNY1320
1321 DNCPDIIEQS EKDNITNHGE AELTLQCVEG MLLSGVPCED IGVMTLYRAQ LRLLKKIFNK1380
1381 NVYDGLEILT ADQFQGRDKK CIIISMVRRN SQLNGGALLK ELRRVNVAMT RAKSKLIIIG1440
1441 SKSTIGSVPE IKSFVNLLEE RNWVYTMCKD ALYKYKFPDR SNAIDEARKG CGKRTGAKPI1500
1501 TSKSKFVSDK PIIKEILQEY ES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
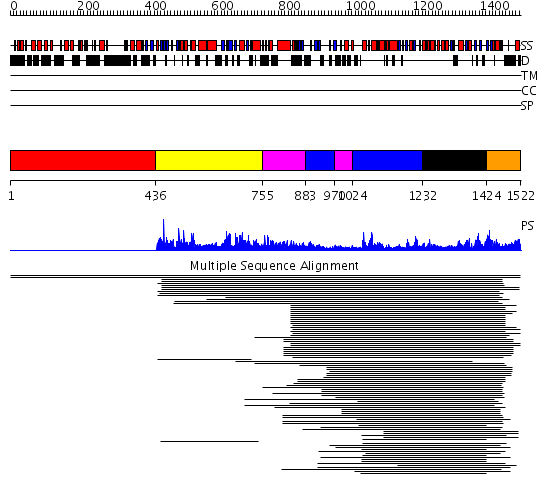
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..435] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [436..754] | 2.021937 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [755..882] [970..1023] |
9.221849 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [883..969] [1024..1231] |
9.221849 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 5 | View Details | [1232..1423] | 64.69897 | DEXX box DNA helicase | |
| 6 | View Details | [1424..1522] | 2.39794 | STRUCTURE OF DNA HELICASE WITH ADPNP |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)