













| General Information: |
|
| Name(s) found: |
DGR2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 852 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFKSKTSTLS YDETPNSNEG DRNATPVNPK EKSQTKHLNI PGDRSRHSSI ADSKRSSSRY 60
61 DGGYSADIIP AQLRFIDNID YGTRLRKTLH RNSVVSNGYN KLSENDRWYF DLFDRKYFEN 120
121 YLEEPTYIKI FKKKEGLEQF DRMFLAQELK IPDVYKSTTY QGEPAVANSE LFKNSICCCT 180
181 FSHDGKYMVI GCKDGSLHLW KVINSPVKRS EMGRSEKSVS ASRANSLKIQ RHLASISSHN 240
241 GSISSNDLKP SDQFEGPSKQ LHLYAPVFYS DVFRVFMEHA LDILDANWSK NGFLITASMD 300
301 KTAKLWHPER KYSLKTFVHP DFVTSAIFFP NDDRFIITGC LDHRCRLWSI LDNEVSYAFD 360
361 CKDLITSLTL SPPGGEYTII GTFNGYIYVL LTHGLKFVSS FHVSDKSTQG TTKNSFHPSS 420
421 EYGKVQHGPR ITGLQCFFSK VDKNLRLIVT TNDSKIQIFD LNEKKPLELF KGFQSGSSRH 480
481 RGQFLMMKNE PVVFTGSDDH WFYTWKMQSF NLSAEMNCTA PHRKKRLSGS MSLKGLLRIV 540
541 SNKSTNDECL TETSNQSSSH TFTNSSKNVL QTQTVGSQAI KNNHYISFHA HNSPVTCASI 600
601 APDVAIKNLS LSNDLIFELT SQYFKEMGQN YSESKETCDN KPNHPVTETG GFSSNLSNVV 660
661 NNVGTILITT DSQGLIRVFR TDILPEIRKK IIEKFHEYNL FHLEAAGKIN NHNNDSILEN 720
721 RMDERSSTED NEFSTTPPSN THNSRPSHDF CELHPNNSPV ISGMPSRASA IFKNSIFNKS 780
781 NGSFISLKSR SESTSSTVFG PHDIPRVSTT YPKLKCDVCN GSNFECASKN PIAGGDSGFT 840
841 CADCGTILNN FR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
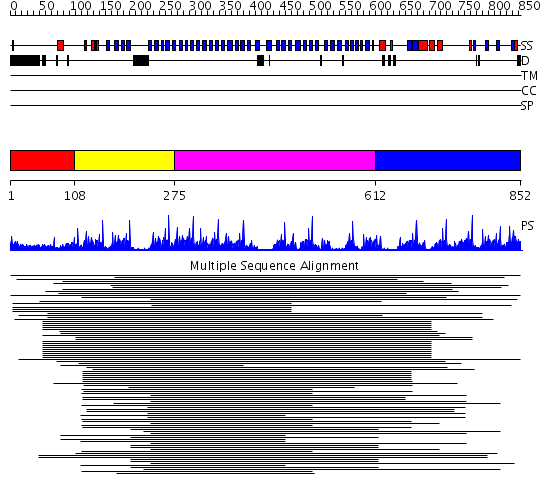
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [108..274] | 126.28043 | Tup1, C-terminal domain | |
| 3 | View Details | [275..611] | 34.221849 | No description for 1scgB_ was found. | |
| 4 | View Details | [612..852] | 2.69897 | TolB, C-terminal domain; TolB, N-terminal domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)