













| General Information: |
|
| Name(s) found: |
DCP2_YEAST
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 970 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLPLRHALE NVTSVDRILE DLLVRFIINC PNEDLSSVER ELFHFEEASW FYTDFIKLMN 60
61 PTLPSLKIKS FAQLIIKLCP LVWKWDIRVD EALQQFSKYK KSIPVRGAAI FNENLSKILL 120
121 VQGTESDSWS FPRGKISKDE NDIDCCIREV KEEIGFDLTD YIDDNQFIER NIQGKNYKIF 180
181 LISGVSEVFN FKPQVRNEID KIEWFDFKKI SKTMYKSNIK YYLINSMMRP LSMWLRHQRQ 240
241 IKNEDQLKSY AEEQLKLLLG ITKEEQIDPG RELLNMLHTA VQANSNNNAV SNGQVPSSQE 300
301 LQHLKEQSGE HNQQKDQQSS FSSQQQPSIF PSLSEPFANN KNVIPPTMPM ANVFMSNPQL 360
361 FATMNGQPFA PFPFMLPLTN NSNSANPIPT PVPPNFNAPP NPMAFGVPNM HNLSGPAVSQ 420
421 PFSLPPAPLP RDSGYSSSSP GQLLDILNSK KPDSNVQSSK KPKLKILQRG TDLNSIKQNN 480
481 NDETAHSNSQ ALLDLLKKPT SSQKIHASKP DTSFLPNDSV SGIQDAEYED FESSSDEEVE 540
541 TARDERNSLN VDIGVNVMPS EKDSRRSQKE KPRNDASKTN LNASAESNSV EWGPGKSSPS 600
601 TQSKQNSSVG MQNKYRQEIH IGDSDAYEVF ESSSDEEDGK KLEELEQTQD NSKLISQDIL 660
661 KENNFQDGEV PHRDMPTESN KSINETVGLS STTNTVKKVP KVKILKRGET FASLANDKKA 720
721 FDSSSNVSSS KDLLQMLRNP ISSTVSSNQQ SPKSQHLSGD EEIMMMLKRN SVSKPQNSEE 780
781 NASTSSINDA NASELLGMLK QKEKDITAPK QPYNVDSYSQ KNSAKGLLNI LKKNDSTGYP 840
841 RTEGGPSSEM STSMKRNDAT NNQELDKNST ELLNYLKPKP LNDGYENISN KDSSHELLNI 900
901 LHGNKNSSAF NNNVYATDGY SLASDNNENS SNKLLNMLQN RSSAINEPNF DVRSNGTSGS 960
961 NELLSILHRK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
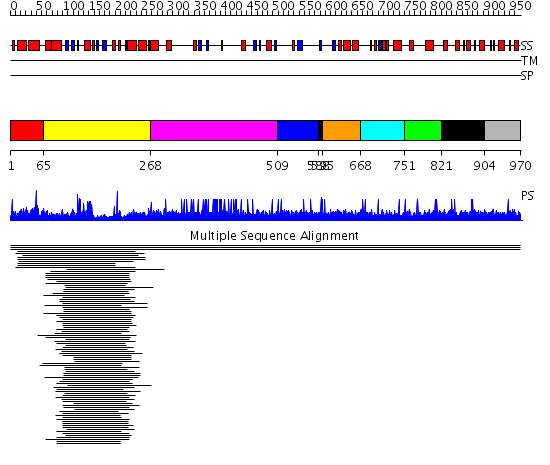
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..64] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [65..267] | 19.0 | Nucleoside triphosphate pyrophosphorylase (MutT) | |
| 3 | View Details | [268..508] | 11.75 | Fibrinogen | |
| 4 | View Details | [509..587] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [588..594] [821..903] |
8.74 | No description for 1l8mA was found. | |
| 6 | View Details | [595..667] | 8.74 | No description for 1l8mA was found. | |
| 7 | View Details | [668..750] | 8.74 | No description for 1l8mA was found. | |
| 8 | View Details | [751..820] | 8.74 | No description for 1l8mA was found. | |
| 9 | View Details | [904..970] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)