













| General Information: |
|
| Name(s) found: |
CTF3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 733 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLILDDIIL SLTNANERTP PQALKTTLSL LYEKSKQYGL SSPQLQALVR LLCETSIIDT 60
61 VTKVYIVENC FLPDGYLTKE LLLEIINHLG TPTVFSRYRI QTPPVLQSAL CKWLVHVYFL 120
121 FPVHSEREHN ISSSIWLHLW QFSFLQKWIT PLVIWQATTP VDVKPWKLSI IKRCAMHPGY 180
181 RDAPGSATLI LQRFQCLVGA SSQITESIIT INCNRKTLKS HRNLKLDAHF LSILKRILSR 240
241 AHPANFPADT VQNTIDMYLS EIHQLGADSI YPLRLQSLPE YVPSDSTVSL WDVTSLEQLA 300
301 QNWPQLHIPN DVDYMMKPSL NSNVLLPRKV MSRDSLKHLY SSIILIKNSR DESSSPYEWC 360
361 IWQLKRCFAH QIETPQEVIP IIISVSSMDN KLSSRIIQTF CNLKYLKLDE LTLKKVCGGI 420
421 LPLWKPELIS GTREFFVKFM ASIFMWSTRD GHDNNCTFSE TCFYVLQMIT NWVLDDKLIA 480
481 LGLTLLHDMQ SLLTLDKIFN NATSNRFSTM AFISSLDILT QLSKQTKSDY AIQYLIVGPD 540
541 IMNKVFSSDD PLLLSAACRY LVATKNKLMQ YPSTNKFVRM QNQYIMDLTN YLYRNKVLSS 600
601 KSLFGVSPDF FKQILENLYI PTADFKNAKF FTITGIPALS YICIIILRRL ETAENTKIKF 660
661 TSGIINEETF NNFFRVHHDE IGQHGWIKGV NNIHDLRVKI LMHLSNTANP YRDIAAFLFT 720
721 YLKSLSKYSV QNS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
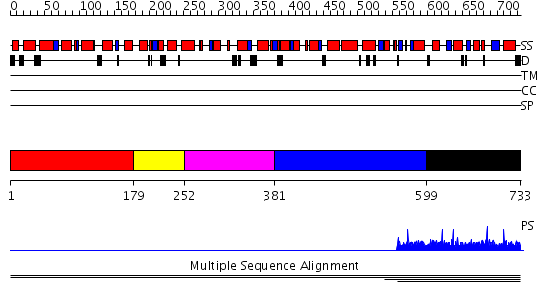
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..178] | 2.1 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 2 | View Details | [179..251] | 2.1 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 3 | View Details | [252..380] | 2.1 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 4 | View Details | [381..598] | 2.1 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 5 | View Details | [599..733] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)