













| General Information: |
|
| Name(s) found: |
BRO1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 844 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKPYLFDLKL KDTEKLDWKK GLSSYLKKSY GSSQWRTFYD EKATSELDHL RNNANGELAP 60
61 SSLSEQNLKY YSFLEHLYFR LGSKGSRLKM DFTWYDAEYS SAQKGLKYTQ HTLAFEKSCT 120
121 LFNIAVIFTQ IARENINEDY KNSIANLTKA FSCFEYLSEN FLNSPSVDLQ SENTRFLANI 180
181 CHAEAQELFV LKLLNDQISS KQYTLISKLS RATCNLFQKC HDFMKEIDDD VAIYGEPKWK 240
241 TTVTCKLHFY KSLSAYYHGL HLEEENRVGE AIAFLDFSMQ QLISSLPFKT WLVEFIDFDG 300
301 FKETLEKKQK ELIKDNDFIY HESVPAVVQV DSIKALDAIK SPTWEKILEP YMQDVANKCD 360
361 SLYRGIIPLD VYEKESIYSE EKATLLRKQV EETETANLEY SSFIEFTNLP RLLSDLEKQF 420
421 SDGNIFSNTD TQGQLMRDQI QTWCKFIQTN EFRDIEEQMN KIVFKRKQIL EILSALPNDQ 480
481 KENVTKLKSS LVAASNSDEK LFACVKPHIV EINLLNDNGK IWKKFDEFNR NTPPQPSLLD 540
541 IDDTKNDKIL ELLKQVKGHA EDLRTLKEER SRNLSELRDE INNDDITKLL IINKGKSDVE 600
601 LKDLFEVELE KFEPLSTRIE ATIYKQSSMI DDIKAKLDEI FHLSNFKDKS SGEEKFLEDR 660
661 KNFFDKLQEA VKSFSIFASD LPKGIEFYDS LFNMSRDLAE RVRVAKQTED STANSPAPPL 720
721 PPLDSKASVV GGPPLLPQKS AAFQSLSRQG LNLGDQFQNL KISAGSDLPQ GPGIPPRTYE 780
781 ASPYAATPTM AAPPVPPKQS QEDMYDLRRR KAVENEEREL QENPTSFYNR PSVFDENMYS 840
841 KYSS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
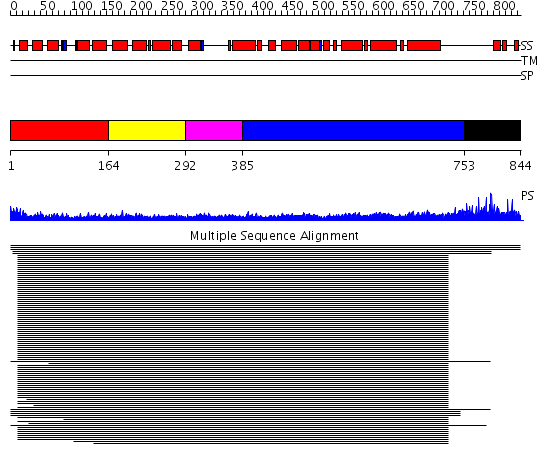
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..163] | 66.148742 | BRO1-like domain No confident structure predictions are available. | |
| 2 | View Details | [164..291] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [292..384] | 6.154902 | Tropomyosin | |
| 4 | View Details | [385..752] | 14.30103 | Heavy meromyosin subfragment | |
| 5 | View Details | [753..844] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)