













| General Information: |
|
| Name(s) found: |
ASE1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 885 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METATSSPLP IKSRRNSENS GSTTVIPHMN PSLATPLTVS TMVNQSNSKE FMKLTPVRIR 60
61 DFGSPLKNVS TNYHFLDSEN GKGNTMDNMY RENFILISKD LEKLLENLNV IYQNIGYSNT 120
121 EIITKEKIIF TTISNSIKQF FEQADEELKR LSAENGIEQD ILNNILERIN DPSGIKTIPD 180
181 LYIRNAILLQ ESKTVPQSPK KPLSLLSKKA ALDTAKKFVL GSFLPRLRDY LKSLITLKHL 240
241 IQSVKENLPG LTEADNEAIA EFPELSTLTA YLLQIENGKG DIGLSMKFII DNRKDILKGS 300
301 AFKTINEESV KHMNEVIKIY EEEYERRFKS VLTKKVSISS ICEQLGTPLA TLIGEDFEQD 360
361 LRSYGEEENS TSEIPNFHPV DRERMSKIDI TLEKLQAIHK ERADKKRLLM EQCQKLWTRL 420
421 KISQEYIKTF MRNNSSLSTE SLGRISKEVM RLEAMKKKLI KKLISDSWDK IQELWRTLQY 480
481 SEESRSKFII VFEELRNSAT TLQEDELLLE TCENELKRLE EKLTLYKPIL KLISDFESLQ 540
541 EDQEFLERSS KDSSRLLSRN SHKILLTEEK MRKRITRHFP RVINDLRIKL EEADGLFDQP 600
601 FLFKGKPLSE AIDIQQQEIE AKYPRCRVRM QRSKKGKCGA NKENKVIKNT FKATESSIRV 660
661 PIGLNLNDAN ITYKTPSKKT IQGLTKNDLS QENSLARHMQ GTTKLSSPNR RATRLLAPTV 720
721 ISRNSKGNIE RPTLNRNRSS DLSSSPRINH THGEHAVKPR QLFPIPLNKV DTKGSHIPQL 780
781 TKEKALELLK RSTGTTGKEN VRSPERKSSL EDYAQKLSSP YKEPEHSIYK LSMSPEGKFQ 840
841 LNIQQKDIES GFDDTSMMED ENDKDFITWK NEQVSKLNGF SFTDI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
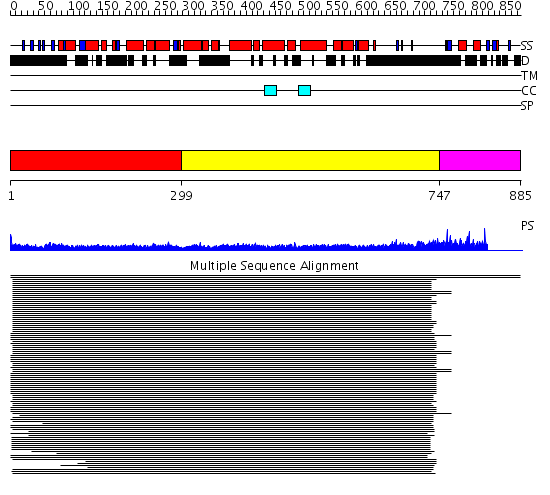
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..298] | 34.365997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [299..746] | 13.154902 | Heavy meromyosin subfragment | |
| 3 | View Details | [747..885] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [737..885] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)