













| General Information: |
|
| Name(s) found: |
ART5_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 586 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSLSSLSSS GGHSEQKERE RISYFDIRIN SPYKDIILIQ GSPLELSSIP LSGNLVISVK 60
61 NEIVVKKISL RLVGRFKLEF LQVGRYKKNS SSLASLVKEK RKIFECYWDN LLVSSKGDVL 120
121 VGGENAENQH NSSSGRSTSN QDMDTSGNAI FLSKRSLSSP VFNKIIRRKT HSSHRKILEL 180
181 PENGVTGTPF EGLRENARSR SSSSNTLNNN SHSYSNRDGS GSSYLFLMKR GNYELPFNTM 240
241 LPPEVCETIE GLQSGSILYS FEAIIDGRQL WDTDLSVHTS PHGPIGSTST SGNGMRTKNK 300
301 IIIKKFKYLR ILRTLSMDNL AMQEEISVGN TWRDKLQYET SIPSRAVPIG STTPVKIKIF 360
361 PFEKNIRLDR IEMALIQYYA MKDSSAQIYD DEIAVMKITH LADFGPLTDK LDVDCPFTIP 420
421 DNLKQITQDC CLQDNLIRVM HKLQVRILLQ RQVDGEYKNL EIKAQLPMLL FISPHLPMKG 480
481 RLVLFDKHDG KIHFRPGELV PLFLTTYPAQ GLTPGVELNS TTTAHLALPQ PPPNYHESTN 540
541 DHLMPALQPL GADSVVLTVP SYEQAQAQAS ASSYVTGSVP AYCDDD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
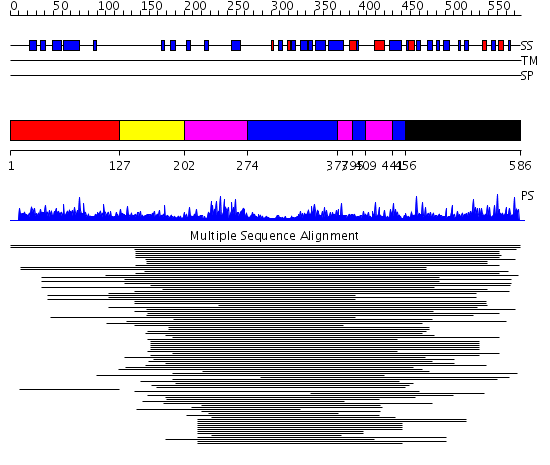
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | 1.03897 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [127..201] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [202..273] [377..394] [409..440] |
16.522879 | Arrestin | |
| 4 | View Details | [274..376] [395..408] [441..455] |
16.522879 | Arrestin | |
| 5 | View Details | [456..586] | 1.068996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
|||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.70 |
Source: Reynolds et al. (2008)