













| General Information: |
|
| Name(s) found: |
AP1B1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 726 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPPLDKRIKK FLKDSIRIAP KISGKGELSE LRTGLVSQYP QTRKDAIKKT IQQMTLGKDV 60
61 SSLFPDVLKN IATIDVEQKK LVYLYVMNYA ETHPELCILA VNTFITDAQD PNPLIRCMAI 120
121 RTMSMIRVDK ILEYIETPLR RTLHDDNAYV RKTAVICVAK LFQLNKDLCV ELGVVEDLVN 180
181 ALDDSNPLVI ANATAALIEI HNMDMDAVDL SSLIQSHVSQ FLLALNECTE WARIIILGTL 240
241 SEYSAKDSLE AQDIIDRVTA HLQHVNPAVV LATIKVIVRN LPQIEYSSNS LIMKRLSSAF 300
301 VSLMSTPPEM QYVALKNIRI ILEKYPELLT KELRIFYVKF NDPLYVKLEK IDILVRLVDP 360
361 SNLKQCTLLL TELKEYAMEY EPEFVSRAIQ ALSQLGIKYA QESFVSKVLD ILLELLERQD 420
421 TIKDDCCISL CDLLRHCPGN DKMAKQVCAV FNTWSNPEVL LQSDIAKCNY VWLLGQHPNN 480
481 FSDLESKINI FIENFVQEEA LTQMSLLMTI VRLHATLTGS MLQSVLELAT QQTHELDVRD 540
541 MAMMYWRCLS MPNNESLVND LCQNKLPMIS NTLEKFSPEV LEKLLMELGT ISSIYFKPDS 600
601 NRRKGKKYVQ NIVKGKHIEE LESMAKNEIS SKANDDVLLD FDERDDVTNT NAGMLNTLTT 660
661 LGDLDDLFDF GPSEDATQIN TNDTKAVQGL KELKLGGDSN GISSGGKNNP DVSGGNIVSQ 720
721 DLLDLF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
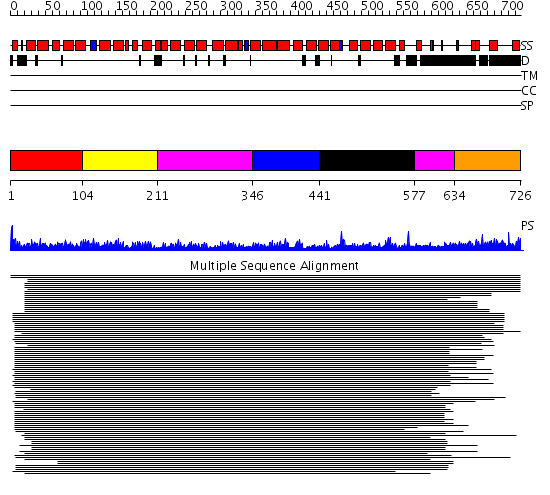
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..103] | 1164.0 | Adaptin beta subunit N-terminal fragment | |
| 2 | View Details | [104..210] | 1164.0 | Adaptin beta subunit N-terminal fragment | |
| 3 | View Details | [211..345] [577..633] |
1164.0 | Adaptin beta subunit N-terminal fragment | |
| 4 | View Details | [346..440] | 1164.0 | Adaptin beta subunit N-terminal fragment | |
| 5 | View Details | [441..576] | 1164.0 | Adaptin beta subunit N-terminal fragment | |
| 6 | View Details | [634..726] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)