













| General Information: |
|
| Name(s) found: |
ALP1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 573 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDETVNIQMS KEGQYEINSS SIIKEEEFVD EQYSGENVTK AITTERKVED DAAKETESSP 60
61 QERREVKRKL KQRHIGMIAL GGTIGTGLII GIGPPLAHAG PVGALISYLF MGTVIYSVTQ 120
121 SLGEMVTFIP VTSSFSVFAQ RFLSPALGAT NGYMYWLSWC FTFALELSVL GKVIQYWTEA 180
181 VPLAAWIVIF WCLLTSMNMF PVKYYGEFEF CIASIKVIAL LGFIIFSFCV VCGAGQSDGP 240
241 IGFRYWRNPG AWGPGIISSD KNEGRFLGWV SSLINAAFTY QGTELVGITA GEAANPRKAL 300
301 PRAIKKVVVR ILVFYILSLF FIGLLVPYND PKLDSDGIFV SSSPFMISIE NSGTKVLPDI 360
361 FNAVVLITIL SAGNSNVYIG SRVLYSLSKN SLAPRFLSNV TRGGVPYFSV LSTSVFGFLA 420
421 FLEVSAGSGK AFNWLLNITG VAGFFAWLLI SFSHIRFMQA IRKRGISRDD LPYKAQMMPF 480
481 LAYYASFFIA LIVLIQGFTA FAPTFQPIDF VAAYISIFLF LAIWLSFQVW FKCRLLWKLQ 540
541 DIDIDSDRRQ IEELVWIEPE CKTRWQRVWD VLS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
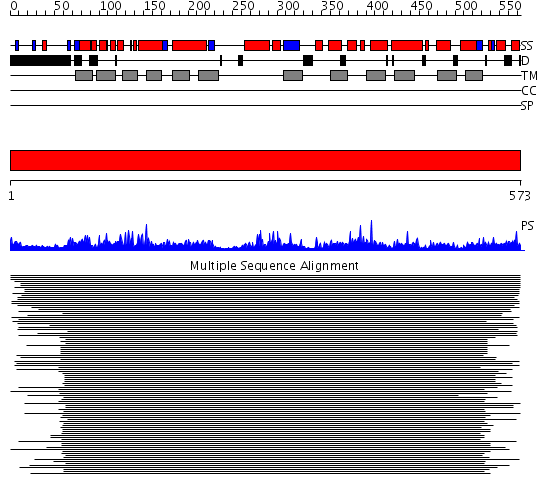
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..573] | 267.013228 | Amino acid permease No confident structure predictions are available. | |
| 2 | View Details | [65..573] | 1.57 | Crystal structure of LEUTAA, a bacterial homolog of Na+/Cl--dependent neurotransmitter transporters |
| Source: Reynolds et al. 2008. Manuscript submitted |

|