













| General Information: |
|
| Name(s) found: |
CPL2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 770 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNRLGHKSVV YHGDLRLGEL DVNHVSSSHE FRFPNDEIRI HHLSPAGERC PPLAILQTIA 60
61 SFAVRCKLES SAPVKSQELM HLHAVCFHEL KTAVVMLGDE EIHLVAMPSK EKKFPCFWCF 120
121 SVPSGLYDSC LRMLNTRCLS IVFDLDETLI VANTMKSFED RIEALKSWIS REMDPVRING 180
181 MSAELKRYMD DRMLLKQYID NDYAFDNGVL LKAQPEEVRP TSDGQEKVCR PVIRLPEKNT 240
241 VLTRIKPEIR DTSVLVKLRP AWEELRSYLT AKTRKRFEVY VCTMAERDYA LEMWRLLDPE 300
301 AHLISLKELR DRIVCVKPDA KKSLLSVFNG GICHPKMAMV IDDRMKVWED KDQPRVHVVS 360
361 AYLPYYAPQA ETALVVPHLC VARNVACNVR GYFFKEFDES LMSSISLVYY EDDVENLPPS 420
421 PDVSNYVVIE DPGFASNGNI NAPPINEGMC GGEVERRLNQ AAAADHSTLP ATSNAEQKPE 480
481 TPKPQIAVIP NNASTATAAA LLPSHKPSLL GAPRRDGFTF SDGGRPLMMR PGVDIRNQNF 540
541 NQPPILAKIP MQPPSSSMHS PGGWLVDDEN RPSFPGRPSG LYPSQFPHGT PGSAPVGPFA 600
601 HPSHLRSEEV AMDDDLKRQN PSRQTTEGGI SQNHLVSNGR EHHTDGGKSN GGQSHLFVSA 660
661 LQEIGRRCGS KVEFRTVIST NKELQFSVEV LFTGEKIGIG MAKTKKDAHQ QAAENALRSL 720
721 AEKYVAHVAP LARETEKGPE NDNGFLWESS EDVSNKGLEE EAPKENISEL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
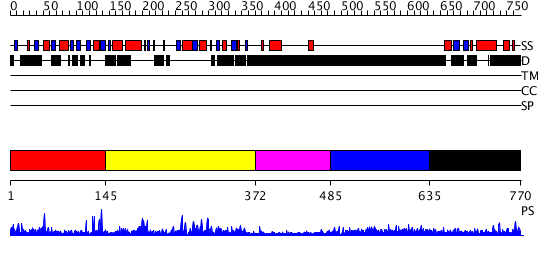
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [145..371] | 16.09691 | Three-dimensional structure of a RNA-polymerase II binding protein with associated ligand. | |
| 3 | View Details | [372..484] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [485..634] | 2.226997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [635..770] | 2.198997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)