













| General Information: |
|
| Name(s) found: |
URH2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 322 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
UDP-glucosyltransferase activity
[IDA]
hydrolase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAIGDRKKII IDTDPGIDDA MAIFVALNSP EVDVIGLTTI FGNVYTTLAT RNALHLLEVA 60
61 GRTDIPVAEG THKTFLNDTK LRIADFVHGK DGLGNQNFPP PKGKPIEKSG PEFLVEQAKL 120
121 CPGEITVVAL GPLTNLALAV QLDPEFSKNV GQIVLLGGAF AVNGNVNPAS EANIFGDPEA 180
181 ADIVFTCGAD IIAVGINVTH QVIMTADDKD KLASSKGKLA QYLCKILDVY YDYHLTAYEI 240
241 KGVYLHDPAT ILAAFLPSLF TYTEGVARVQ TSGITRGLTL LYNNLKRFEE ANEWSDKPTV 300
301 KVAVTVDAPA VVKLIMDRLM ES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
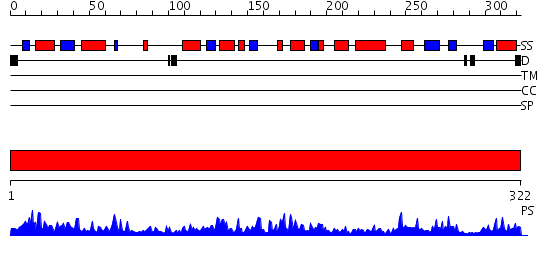
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..322] | 102.0 | Crystal structure of a the E. coli pyrimidine nucleoside hydrolase YbeK with bound ribose |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)