













| General Information: |
|
| Name(s) found: |
NADC_YEAST
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 295 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPVYEHLLPV NGAWRQDVTN WLSEDVPSFD FGGYVVGSDL KEANLYCKQD GMLCGVPFAQ 60
61 EVFNQCELQV EWLFKEGSFL EPSKNDSGKI VVAKITGPAK NILLAERTAL NILSRSSGIA 120
121 TASHKIISLA RSTGYKGTIA GTRKTTPGLR RLEKYSMLVG GCDTHRYDLS SMVMLKDNHI 180
181 WATGSITNAV KNARAVCGFA VKIEVECLSE DEATEAIEAG ADVIMLDNFK GDGLKMCAQS 240
241 LKNKWNGKKH FLLECSGGLN LDNLEEYLCD DIDIYSTSSI HQGTPVIDFS LKLAH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
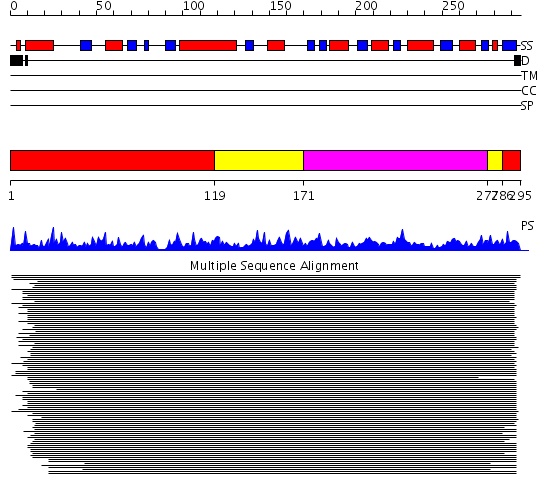
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] [286..295] |
398.69897 | Quinolinic acid phosphoribosyltransferase (Nicotinate-nucleotide pyrophosphorylase, NadC), C-terminal domain; Quinolinic acid phosphoribosyltransferase (Nicotinate-nucleotide pyrophosphorylase, NadC), N-terminal domain | |
| 2 | View Details | [119..170] [277..285] |
398.69897 | Quinolinic acid phosphoribosyltransferase (Nicotinate-nucleotide pyrophosphorylase, NadC), C-terminal domain; Quinolinic acid phosphoribosyltransferase (Nicotinate-nucleotide pyrophosphorylase, NadC), N-terminal domain | |
| 3 | View Details | [171..276] | 398.69897 | Quinolinic acid phosphoribosyltransferase (Nicotinate-nucleotide pyrophosphorylase, NadC), C-terminal domain; Quinolinic acid phosphoribosyltransferase (Nicotinate-nucleotide pyrophosphorylase, NadC), N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)