













| General Information: |
|
| Name(s) found: |
ALG1_YEAST
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 449 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFLEIPRWLL ALIILYLSIP LVVYYVIPYL FYGNKSTKKR IIIFVLGDVG HSPRICYHAI 60
61 SFSKLGWQVE LCGYVEDTLP KIISSDPNIT VHHMSNLKRK GGGTSVIFMV KKVLFQVLSI 120
121 FKLLWELRGS DYILVQNPPS IPILPIAVLY KLTGCKLIID WHNLAYSILQ LKFKGNFYHP 180
181 LVLISYMVEM IFSKFADYNL TVTEAMRKYL IQSFHLNPKR CAVLYDRPAS QFQPLAGDIS 240
241 RQKALTTKAF IKNYIRDDFD TEKGDKIIVT STSFTPDEDI GILLGALKIY ENSYVKFDSS 300
301 LPKILCFITG KGPLKEKYMK QVEEYDWKRC QIEFVWLSAE DYPKLLQLCD YGVSLHTSSS 360
361 GLDLPMKILD MFGSGLPVIA MNYPVLDELV QHNVNGLKFV DRRELHESLI FAMKDADLYQ 420
421 KLKKNVTQEA ENRWQSNWER TMRDLKLIH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
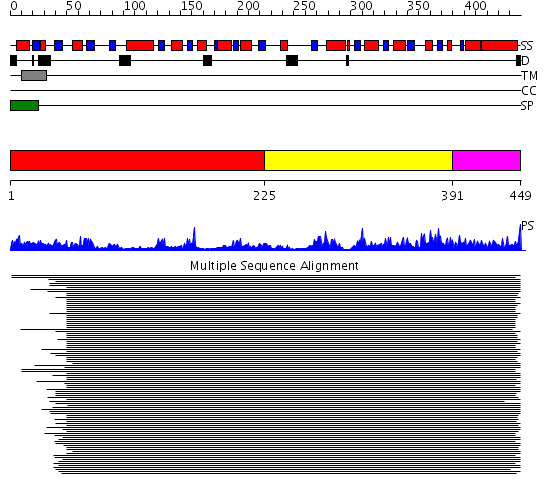
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..224] | 6.69897 | Peptidoglycan biosynthesys glycosyltransferase MurG | |
| 2 | View Details | [225..390] | 6.69897 | Peptidoglycan biosynthesys glycosyltransferase MurG | |
| 3 | View Details | [391..449] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|