













| General Information: |
|
| Name(s) found: |
nuo-2 /
CE37406
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 445 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA]
|
| Biological Process: |
electron transport
[IEA mitochondrial electron transport, NADH to ubiquinone [IEA nematode larval development [IMP determination of adult lifespan [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP growth [IMP |
| Molecular Function: |
NADH dehydrogenase (quinone) activity
[IEA RNA binding [IEA metal cluster binding [IEA NADH dehydrogenase (ubiquinone) activity [IEA oxidoreductase activity, acting on NADH or NADPH [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSEILNYPI HRFLDSSLLG QFIGPEGSNI YKIEKYNKVA LDIWKNDEEN SNVRITGPYW 60
61 NLKSALNDVL ELVSTIRNKN QRYKFEMPSK DIGFLIGKNG AKINEIKLSS NVDVHFERNN 120
121 ENRDNGETDG RVVVSVTGNY QQILIGLRLI YDRLTSKGQK TLYEDPRTRQ FAESLMKMLG 180
181 SVIRQAVSRQ IVRNSPISTT AAVAQTNQTG DKKESPKKPT IWKIDEHKRE RLANFGKYAA 240
241 ECLPKFVQKV QFAAGDELEL LIHPSGVVPV LSFLKGNHSA QFTNLTFITG MDVPTRKNRL 300
301 EVIYSLYSVR FNARVRVRTY TDEIAPIDSA TPVFKGADWF EREVYDMYGV WFNNHPDLRR 360
361 ILTDYGFEGH PFRKDYPLSG YNEVRYDPEL KRVVYEPSEL AQEFRKFDLN TPWETFPAFR 420
421 NQSITSGYET ILEVAEPTPA TPQNK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
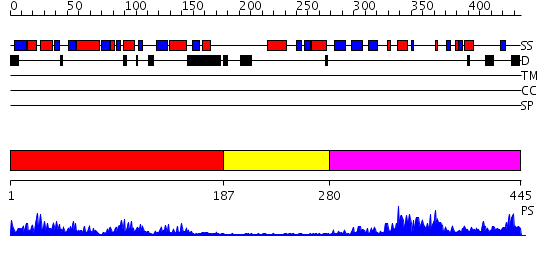
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..186] | 32.09691 | No description for 2annA was found. | |
| 2 | View Details | [187..279] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [280..445] | 27.09691 | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)