













| General Information: |
|
| Name(s) found: |
PTN2B_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 632 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein amino acid dephosphorylation
[IEA]
dephosphorylation [IEA] |
| Molecular Function: |
protein tyrosine phosphatase activity
[IEA]
phosphatase activity [IEA] protein tyrosine/serine/threonine phosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METDPANSSS KSPAVVSEKD VLIPEPSENT VGVVQDPVSA EREAHEDSIS TEASVAKVDD 60
61 TQMPASSTGS EPLSKTDDIV PCPPGSSPRE SPPSIFSSSG LSSWAKSFKF QQQDPNRTDS 120
121 GMSAFTRFTS ELGLHLPTKG SEEVGDSRSS NTQVGGAFES LTKAVVDSSR GAVKAMQVKA 180
181 RHIVSQNKRR YQEGEFDLDM TYITENIIAM GFPAGDISSG LFGFFEGLYR NHMEEVIKFF 240
241 ETHHKDKYKV YNLCSERLYD ASRFEGKVAS FPFDDHNCPP IQLIPSFCQS AYTWLKEDIQ 300
301 NVVVVHCKAG MARTGLMICC LLLYLKFFPT AEEAIDYYNQ KRCLDGKALV LPSQIRYVKY 360
361 YERVQNQFDG KVPPERRCML RGFRLINCPY WIRPAITISN HTDILFSTKK HQKTKDLGPE 420
421 DFWIKAPKKG VVVFAIPGEA GLTELAGDFK IHFQDSDGDF YCWLNTTLTD NRTMLKGSDF 480
481 DGFDKRKLPA PGFHVEIVMI EPDNSQPTKS KSDSTQQQSQ SSSSADSSKL KSNEKDDDVF 540
541 SDSDGEEEGN SQSYSTNEKT ASSMHTTSKP HQINEPPKRD DPSANRSVTS SSSSGHYNPI 600
601 PNNSLAVSDI KAIAADASVF SFGDEEEDYE SD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
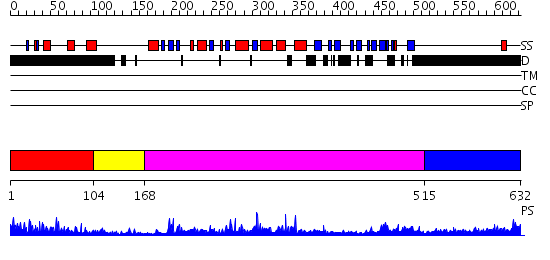
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..103] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [104..167] | 51.09691 | Tyrosine phosphatase | |
| 3 | View Details | [168..514] | 74.30103 | Pten tumor suppressor (Phoshphoinositide phosphatase), C-terminal domain; Phoshphoinositide phosphatase Pten (Pten tumor suppressor), N-terminal domain | |
| 4 | View Details | [515..632] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)