













| General Information: |
|
| Name(s) found: |
FREE1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 601 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
signal transduction
[ISS]
|
| Molecular Function: |
phosphoinositide binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQQGDYNSYY HHQYSQFQNP TPNPNPNPNP SPPAPATVAG PTDLTRNTYA SAPPFTGGYG 60
61 SADYSNYSQN YTPYGQNSEH VPPSAPSFTS PSQPPPSPPA TSLNPNSYST FNQPPPPPTI 120
121 HPQPLSSYGS FDSTAPYQQP TSQHMYYSPY DQHQTSGYSS APPPSSAPAP NPNPAPYSSS 180
181 LYSAPPYSSG GSSIPPSYEK PSVKFDQSGY DGYNRSRSDL GSDLYGKRSD SGEYPAFEDS 240
241 YGDGVYAYQG GKVEPYGSRG TAPKSSNSTL FDDYGRSISF SSSGRDSSVS SNSAKIVRAV 300
301 PKADVQEDST GGVQKFRVKL LAETYGQTTT DVLCQIGLDG LRMLDPSTSR TLRIYPLENI 360
361 TRCEKLDSSI LAFWSKTPVD IEAKRIRLQS NSYTTNTLLD TVTAAMFQAK EIGGSSRPPT 420
421 SGKLIEQTAE KKKGLGDWMN IIKPVNEEKD HWVPDEAVSK CTSCGSDFGA FIRRHHCRNC 480
481 GDVFCDKCTQ GRIALTAEDN APQVRVCDRC MAEVSQRLSN AKETTGRNVS LQSHEDLARK 540
541 LQEEMERNRK SSSGLREGSG RRMKEVACPT CTVHLQVQVP VSGSETIECG VCQNPFLVSA 600
601 H |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
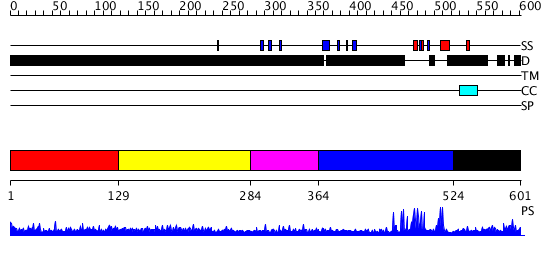
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | 2.159995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [129..283] | 7.177995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [284..363] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [364..523] | 26.09691 | Hrs | |
| 5 | View Details | [524..601] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)