













| General Information: |
|
| Name(s) found: |
NPY1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 571 amino acids |
Gene Ontology: |
|
| Cellular Component: |
late endosome
[IDA]
plasma membrane [IDA] |
| Biological Process: |
inflorescence development
[IGI]
flower development [IGI] apical protein localization [IGI] phyllome development [IGI] response to light stimulus [ISS] basipetal auxin transport [IGI] positive regulation of flower development [IGI] cotyledon development [IGI] |
| Molecular Function: |
protein binding
[IEA]
signal transducer activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKFMKLGSKP DTFESDGKFV KYAVSDLDSD VTIHVGEVTF HLHKFPLLSK SNRMQRLVFE 60
61 ASEEKTDEIT ILDMPGGYKA FEICAKFCYG MTVTLNAYNI TAVRCAAEYL EMTEDADRGN 120
121 LIYKIEVFLN SGIFRSWKDS IIVLQTTRSL LPWSEDLKLV GRCIDSVSAK ILVNPETITW 180
181 SYTFNRKLSG PDKIVEYHRE KREENVIPKD WWVEDVCELE IDMFKRVISV VKSSGRMNNG 240
241 VIAEALRYYV ARWLPESMES LTSEASSNKD LVETVVFLLP KVNRAMSYSS CSFLLKLLKV 300
301 SILVGADETV REDLVENVSL KLHEASVKDL LIHEVELVHR IVDQFMADEK RVSEDDRYKE 360
361 FVLGNGILLS VGRLIDAYLA LNSELTLSSF VELSELVPES ARPIHDGLYK AIDTFMKEHP 420
421 ELTKSEKKRL CGLMDVRKLT NEASTHAAQN ERLPLRVVVQ VLYFEQLRAN HSPVASVAAS 480
481 SHSPVEKTEE NKGEEATKKV ELSKKSRGSK STRSGGGAQL MPSRSRRIFE KIWPGKGEIS 540
541 NKSSEVSSGS SQSPPAKSSS SSSRRRRHSI S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
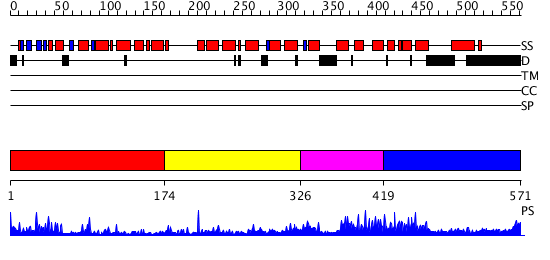
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..173] | 23.221849 | No description for 2if5A was found. | |
| 2 | View Details | [174..325] | 5.546991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [326..418] | 2.166991 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [419..571] | 2.069984 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)