













| General Information: |
|
| Name(s) found: |
RAP24_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 334 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC][IDA]
|
| Biological Process: |
cellular response to salt stress
[IEP]
response to salt stress [IEP] response to osmotic stress [IEP] red or far-red light signaling pathway [IGI] regulation of transcription, DNA-dependent [ISS] ethylene mediated signaling pathway [IMP] response to light stimulus [IEP] response to water deprivation [IEP][IMP] |
| Molecular Function: |
DNA binding
[IDA][TAS]
transcription factor activity [IDA][ISS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAAMNLYTC SRSFQDSGGE LMDALVPFIK SVSDSPSSSS AASASAFLHP SAFSLPPLPG 60
61 YYPDSTFLTQ PFSYGSDLQQ TGSLIGLNNL SSSQIHQIQS QIHHPLPPTH HNNNNSFSNL 120
121 LSPKPLLMKQ SGVAGSCFAY GSGVPSKPTK LYRGVRQRHW GKWVAEIRLP RNRTRLWLGT 180
181 FDTAEEAALA YDKAAYKLRG DFARLNFPNL RHNGSHIGGD FGEYKPLHSS VDAKLEAICK 240
241 SMAETQKQDK STKSSKKREK KVSSPDLSEK VKAEENSVSI GGSPPVTEFE ESTAGSSPLS 300
301 DLTFADPEEP PQWNETFSLE KYPSYEIDWD SILA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
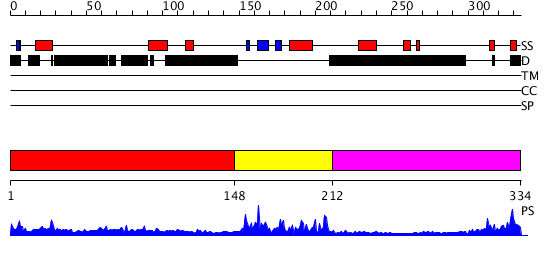
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [148..211] | 22.0 | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | |
| 3 | View Details | [212..334] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)