













| General Information: |
|
| Name(s) found: |
gi|30684462
[NCBI NR]
gi|23297499 [NCBI NR] gi|222422929 [NCBI NR] gi|18401465 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 710 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
DNA methylation
[IEA]
|
| Molecular Function: |
DNA binding
[IEA]
methyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADMRRRNGS GGSSNHERNE QILFPKPETL DFDLPCDTSF PQQIGDNAAS SSGSNVKSLL 60
61 IEMGFCPTLV QKAIDENGQD DFELLLEILT KSTETEPPGP SFHGLMEPKP EPDIEYETDR 120
121 IRIALLTMKF PENLVDFALD RLGKDTPIDE MVDFIVAAQL AEKYAEESED SLDGAEINEE 180
181 DEDVTPVTAR GPEVPNEQLF ETMDKTLRLL EMGFSNDEIS MAIEKIGTKG QISVLAESIV 240
241 TGEFPAECHD DLEDIEKKVS AAAPAVNRTC LSKSWRFVGV GAQKEDGGGG SSSGTANIKP 300
301 DPGIESFPFP ATDNVGETSR GKRPKDEDEN AYPEEYTGYD DRGKRLRPED MGDSSSFMET 360
361 PWMQDEWKDN TYEFPSVMQP RLSQSLGPKV ARRPYFFYGQ LGELSPSWWS KISGFLFGIH 420
421 PEHVDTRLCS ALRRTEGYLH NLPTVNRFNT LPNPRLTIQD AMPHMRSWWP QWDIRKHFNS 480
481 GTCSNMKDAT LLCERIGRRI AECKGKPTQQ DQTLILRHCH TSNLIWIAPN ILSPLEPEHL 540
541 ECIMGYPMNH TNIGGGRLAE RLKLFDYCFQ TDTLGYHLSV LKSMFPQGLT VLSLFSGIGG 600
601 AEIALDRLGI HLKGVVSVES CGLSRNILKR WWQTSGQTGE LVQIEEIKSL TAKRLETLMQ 660
661 RFGGFDFVIC QNPSTPLDLS KEISNSEACE FDYTLFNEFA RVTKRVRDMM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
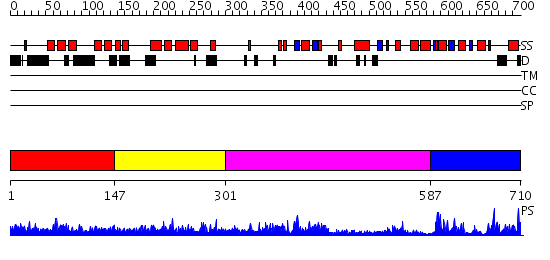
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..146] | 2.14 | Structure of the DNA repair protein hHR23a | |
| 2 | View Details | [147..300] | 71.09691 | No description for 2pv0B was found. | |
| 3 | View Details | [301..586] | 41.0 | Structure of the Q237W mutant of HhaI DNA methyltransferase: an insight into protein-protein interactions | |
| 4 | View Details | [587..710] | 18.69897 | DNMT2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 |
|
|||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)