













| General Information: |
|
| Name(s) found: |
gi|30681531
[NCBI NR]
gi|23296847 [NCBI NR] gi|13430572 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1115 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTMMTPPPVD PEDEEMLVPN SDLVDGPAQP MEVTQPETAA STVENQPAED PPTLKFTWTI 60
61 PNFSRQNTRK HYSDVFVVGG YKWRILIFPK GNNVDHLSMY LDVSDAASLP YGWSRYAQFS 120
121 LAVVNQIHTR YTVRKETQHQ FNARESDWGF TSFMPLSELY DPSRGYLVND TVLVEAEVAV 180
181 RKVLDYWSYD SKKETGFVGL KNQGATCYMN SLLQTLYHIP YFRKAVYHMP TTENDAPTAS 240
241 IPLALQSLFY KLQYNDTSVA TKELTKSFGW DTYDSFMQHD VQELNRVLCE KLEDKMKGTV 300
301 VEGTIQQLFE GHHMNYIECI NVDFKSTRKE SFYDLQLDVK GCKDVYASFD KYVEVERLEG 360
361 DNKYHAEGHG LQDAKKGVLF IDFPPVLQLQ LKRFEYDFMR DTMVKINDRY EFPLELDLDR 420
421 EDGKYLSPDA DRSVRNLYTL HSVLVHSGGV HGGHYYAFIR PTLSDQWYKF DDERVTKEDL 480
481 KRALEEQYGG EEELPQTNPG FNNNPPFKFT KYSNAYMLVY IRESDKDKII CNVDEKDIAE 540
541 HLRVRLKKEQ EEKEDKRRYK AQAHLYTIIK VARDEDLKEQ IGKDIYFDLV DHDKVRSFRI 600
601 QKQTPFQQFK EEVAKEFGVP VQLQRFWIWA KRQNHTYRPN RPLTPQEELQ PVGQIREASN 660
661 KANTAELKLF LEVEHLDLRP IPPPEKSKED ILLFFKLYDP EKAVLSYAGR LMVKSSSKPM 720
721 DITGKLNEMV GFAPDEEIEL FEEIKFEPCV MCEHLDKKTS FRLCQIEDGD IICFQKPLVN 780
781 KEIECLYPAV PSFLEYVQNR QLVRFRALEK PKEDEFVLEL SKQHTYDDVV EKVAEKLGLD 840
841 DPSKLRLTSH NCYSQQPKPQ PIKYRGVDHL SDMLVHYNQT SDILYYEVLD IPLPELQGLK 900
901 TLKVAFHHAT KEEVVIHNIR LPKQSTVGDV INELKTKVEL SHPDAELRLL EVFYHKIYKI 960
961 FPSTERIENI NDQYWTLRAE EIPEEEKNIG PNDRLILVYH FAKETGQNQQ VQNFGEPFFL1020
1021 VIHEGETLEE IKNRIQKKLH VSDEDFAKWK FAFMSMGRPE YLQDTDVVYN RFQRRDVYGA1080
1081 FEQYLGLEHA DTTPKRAYAA NQNRHAYEKP VKIYN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
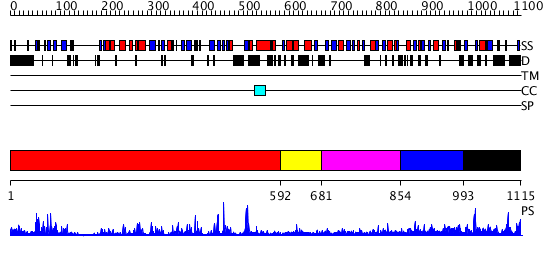
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..591] | 76.154902 | Crystal structure of HAUSP | |
| 2 | View Details | [592..680] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [681..853] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [854..992] | 1.148995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [993..1115] | 1.117977 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)