













| General Information: |
|
| Name(s) found: |
SWI3D_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 985 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
DNA binding
[IEA][ISS]
zinc ion binding [IEA] transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEKRRDSAG TLAFAGSSGD SPASEPMPAP RRRGGGLKRK ANALGGSNFF SSAPSKRMLT 60
61 REKAMLASFS PVHNGPLTRA RQAPSIMPSA ADGVKSEVLN VAVGADGEKP KEEEERNKAI 120
121 REWEALEAKI EADFEAIRSR DSNVHVVPNH CGWFSWEKIH PLEERSLPSF FNGKLEGRTS 180
181 EVYREIRNWI MGKFHSNPNI QIELKDLTEL EVGDSEAKQE VMEFLDYWGL INFHPFPPTD 240
241 TGSTASDHDD LGDKESLLNS LYRFQVDEAC PPLVHKPRFT AQATPSGLFP DPMAADELLK 300
301 QEGPAVEYHC NSCSADCSRK RYHCPKQADF DLCTECFNSG KFSSDMSSSD FILMEPAEAP 360
361 GVGSGKWTDQ ETLLLLEALE IFKENWNEIA EHVATKTKAQ CMLHFLQMPI EDAFLDQIDY 420
421 KDPISKDTTD LAVSKDDNSV LKDAPEEAEN KKRVDEDETM KEVPEPEDGN EEKVSQESSK 480
481 PGDASEETNE MEAEQKTPKL ETAIEERCKD EADENIALKA LTEAFEDVGH SSTPEASFSF 540
541 ADLGNPVMGL AAFLVRLAGS DVATASARAS IKSLHSNSGM LLATRHCYIL EDPPDNKKDP 600
601 TKSKSCSADA EGNDDNSHKD DQPEEKSKKA EEVSLNSDDR EMPDTDTGKE TQDSVSEEKQ 660
661 PGSRTENSTT KLDAVQEKRS SKPVTTDNSE KPVDIICPSQ DKCSGKELQE PLKDGNKLSS 720
721 ENKDASQSTV SQSAADASQP EASRDVEMKD TLQSEKDPED VVKTVGEKVQ LAKEEGANDV 780
781 LSTPDKSVSQ QPIGSASAPE NGTAGGNPNI EGKKEKDICE GTKDKYNIEK LKRAAISAIS 840
841 AAAVKAKNLA KQEEDQIRQL SGSLIEKQLH KLEAKLSIFN EAESLTMRVR EQLERSRQRL 900
901 YHERAQIIAA RLGVPPSMSS KASLPTNRIA ANFANVAQRP PMGMAFPRPP MPRPPGFPVP 960
961 GSFVAATTMT GSSDPSPGSD NVSSV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
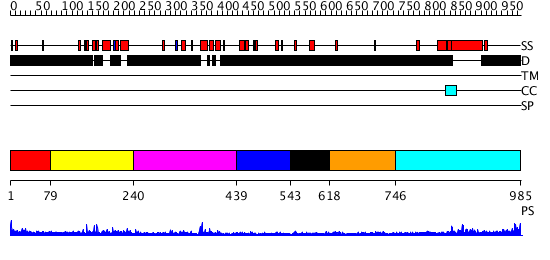
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [79..239] | 13.537602 | No description for PF04433.9 was found. No confident structure predictions are available. | |
| 3 | View Details | [240..438] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [439..542] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [543..617] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [618..745] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [746..985] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.59 |
Source: Reynolds et al. (2008)