













| General Information: |
|
| Name(s) found: |
gi|79313355
[NCBI NR]
gi|22331303 [NCBI NR] gi|17529232 [NCBI NR] gi|110742225 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 950 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
glucosylceramide catabolic process
[IEA]
sphingolipid metabolic process [IEA] |
| Molecular Function: |
glucosylceramidase activity
[IEA]
catalytic activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVGATLFHRR KHSWPTEEFI SRSTLQLLDF DSAAPPPHAW RRRLNCHANI LKEFTITFRE 60
61 AIKMVRLGIR LWSYVREEAS HGRKAPIDPF TKENCKPSAS QGVPLGGMGS GSISRGFRGE 120
121 FKQWQITPGT CDPSPMMSNQ FSIFISRDGG HKKYASVLSP GQHGSLGKSR DKGLSSWGWN 180
181 LNGQHSTYHA LFPRAWTIYD GEPDPELKIS CRQISPFIPN NYRDSSLPAA VFVYTLVNTG 240
241 KERAKVSLLF TWANSMGGTS HMSGGHVNEP FIGEDGVSGV LLHHKTGKGN PPVTFAIAAS 300
301 ETQNVNVTVL PCFGLSEDSS FTAKDMWDTM EQDGKFDQEN FNSGPSTPSL AGDTICAAVS 360
361 ASAWVEAHGK CTVSFALSWS SPKVKFSKGS TYDRRYTKFY GTSPRAALDL VHDALTNYKR 420
421 WEEDIEAWQN PILRDERLPE WYKFTLFNEL YFLVAGGTVW IDSSSLNANG NSQHQQSGLG 480
481 NSDGKVGGLD INDQRNDLGN GNSVGVKSND EVSAIHNRNG LFVDTPHVDD GDDVGRFLYL 540
541 EGVEYVMWCT YDVHFYASYA LLMLFPKIEL NIQRDFAKAV LSEDGRKVKF LAEGNVGIRK 600
601 VRGAVPHDLG MHDPWNEMNA YNIHDTSRWK DLNPKFVLQV YRDFAATGDY QFGIDVWPAV 660
661 RAAMEYMEQF DRDNDDLIEN DGFPDQTYDT WTVHGVSAYC GCLWLAALQA AAAMALQIGD 720
721 KFFAELCKNK FLNAKAALET KLWNGSYFNY DSGSSSNSKS IQTDQLAGQW YAASSGLPPL 780
781 FEESKIRSTM QKIFDFNVMK TKGGKMGAVN GMHPDGKVDD TCMQSREIWT GVTYAAAATM 840
841 ILSGMEEQGF TTAEGIFTAG WSEEGFGYWF QTPEGWTMDG HYRSLIYMRP LAIWGMQWAL 900
901 SLPKAILDAP QINMMDRVHL SPRSRRFSNN FKVVKHKAKC FGNSALSCSC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
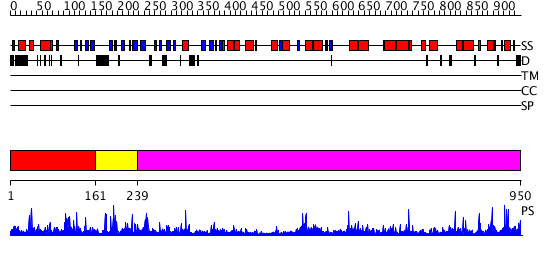
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..160] | 2.039977 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [161..238] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [239..950] | 73.154902 | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)