













| General Information: |
|
| Name(s) found: |
GFA2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 456 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA]
|
| Biological Process: |
synergid death
[IMP]
polar nucleus fusion [IMP] chaperone mediated protein folding requiring cofactor [IGI] embryo sac development [IMP] protein folding [ISS] nuclear membrane fusion [IMP] cellularization of the embryo sac [IMP] |
| Molecular Function: |
heat shock protein binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVPSNGAKVL RLLSRRCLSS SLIQDLANQK LRGVCIGSYR RLNTSVGNHA NVIGDYASKS 60
61 GHDRKWINFG GFNTNFGSTR SFHGTGSSFM SAKDYYSVLG VSKNAQEGEI KKAYYGLAKK 120
121 LHPDMNKDDP EAETKFQEVS KAYEILKDKE KRDLYDQVGH EAFEQNASGG FPNDQGFGGG 180
181 GGGGFNPFDI FGSFNGDIFN MYRQDIGGQD VKVLLDLSFM EAVQGCSKTV TFQTEMACNT 240
241 CGGQGVPPGT KREKCKACNG SGMTSLRRGM LSIQTTCQKC GGAGQTFSSI CKSCRGARVV 300
301 RGQKSVKVTI DPGVDNSDTL KVARVGGADP EGDQPGDLYV TLKVREDPVF RREGSDIHVD 360
361 AVLSVTQAIL GGTIQVPTLT GDVVVKVRPG TQPGHKVVLR NKGIRARKST KFGDQYVHFN 420
421 VSIPANITQR QRELLEEFSK AEQGEYEQRT ATGSSQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
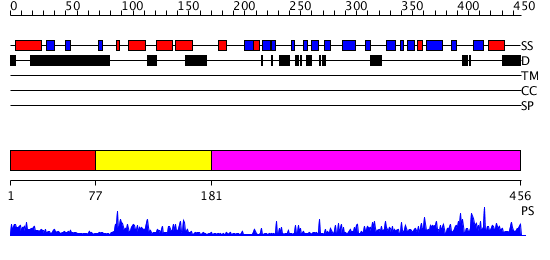
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | 4.304997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [77..180] | 23.30103 | J-DOMAIN (RESIDUES 1-77) OF THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 1-104) OF THE MOLECULAR CHAPERONE DNAJ, NMR, 20 STRUCTURES | |
| 3 | View Details | [181..456] | 41.69897 | The crystal structure of Hsp40 Ydj1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)