













| General Information: |
|
| Name(s) found: |
REV3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1890 amino acids |
Gene Ontology: |
|
| Cellular Component: |
zeta DNA polymerase complex
[ISS]
|
| Biological Process: |
DNA repair
[IDA]
response to UV [IMP] response to UV-B [IMP] |
| Molecular Function: |
DNA binding
[ISS]
DNA-directed DNA polymerase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADSQSGSNV FSLRIVSIDY YMASPIPGYN ICYSSFQGSE VNEVPVIRIY GSTPAGQKTC 60
61 LHIHRALPYL YIPCSEIPLE HHKGVDGSTL ALSLELEKAL KLKGNAASKR QHIHDCEIVR 120
121 AKKFYGYHST EEAFVKIYLS YHPPDVARAA SLLLAGAVLG KSLQPYESHI PFILQFLVDY 180
181 NLYGMGHVHI SKMKFRSPVP HHFRPRRFDL DDCPGQRIDE VAITKANSSA AASVSFPVWS 240
241 LSTIPGQWMW NLSEESDTPL SQSQHRHQHH YRRQSLCELE GDATSSDILN QQFKMYNSLS 300
301 QAQSDTNMVQ SLVAIWEEEY ERTGVHDAPI PPDPGKPSAA DVLQTMSDYV GFGNMLKEML 360
361 NKVELSPPGM KPTAVSSAGP DMHAKPEITD LQALNHMVGT CSEFPASEQL SPLGEKSEEA 420
421 SMENDEYMKT PTDRDTPAQI QDAEALGLFK WFASSQAAED INSDDEILRE TILSPLLPLA 480
481 SINKVLEMAS TDYVSQSQKE CQDILDSQEN LPDFGSSTKR ALPSNPDSQN LRTSSDKQSL 540
541 EIEVASDVPD SSTSNGASEN SFRRYRKSDL HTSEVMEYKN RSFSKSNKPS NSVWGPLPFT 600
601 LTKNLQKDFD STNASDKLGL TKISSYPMNE MTDNYIVPVK EHQADVCNTI DRNVLAGCSL 660
661 RDLMRKKRLC HGESPVSQHM KSRKVRDSRH GEKNECTLRC EAKKQGPALS AEFSEFVCGD 720
721 TPNLSPIDSG NCECNISTES SELHSVDRCS AKETASQNSD EVLRNLSSTT VPFGKDPQTV 780
781 ESGTLVSSNI HVGIEIDSVQ KSGREQESTA NETDETGRLI CLTLSKKPPS LDCLSAGLQD 840
841 SAHSHEIHAR EKQHDEYEGN SNDIPFFPLE DNKEEKKHFF QGTSLGIPLH HLNDGSNLYL 900
901 LTPAFSPPSV DSVLQWISND KGDSNIDSEK QPLRDNHNDR GASFTDLASA SNVVSVSEHV 960
961 EQHNNLFVNS ESNAYTESEI DLKPKGTFLN LNLQASVSQE LSQISGPDGK SGPTPLSQMG1020
1021 FRDPASMGAG QQLTILSIEV HAESRGDLRP DPRFDSVNVI ALVVQNDDSF VAEVFVLLFS1080
1081 PDSIDQRNVD GLSGCKLSVF LEERQLFRYF IETLCKWDPD VLLGWDIQGG SIGFLAERAA1140
1141 QLGIRFLNNI SRTPSPTTTN NSDNKRKLGN NLLPDPLVAN PAQVEEVVIE DEWGRTHASG1200
1201 VHVGGRIVLN AWRLIRGEVK LNMYTIEAVS EAVLRQKVPS IPYKVLTEWF SSGPAGARYR1260
1261 CIEYVIRRAN LNLEIMSQLD MINRTSELAR VFGIDFFSVL SRGSQYRVES MLLRLAHTQN1320
1321 YLAISPGNQQ VASQPAMECV PLVMEPESAF YDDPVIVLDF QSLYPSMIIA YNLCFSTCLG1380
1381 KLAHLKMNTL GVSSYSLDLD VLQDLNQILQ TPNSVMYVPP EVRRGILPRL LEEILSTRIM1440
1441 VKKAMKKLTP SEAVLHRIFN ARQLALKLIA NVTYGYTAAG FSGRMPCAEL ADSIVQCGRS1500
1501 TLEKAISFVN ANDNWNARVV YGDTDSMFVL LKGRTVKEAF VVGQEIASAI TEMNPHPVTL1560
1561 KMEKVYHPCF LLTKKRYVGY SYESPNQREP IFDAKGIETV RRDTCEAVAK TMEQSLRLFF1620
1621 EQKNISKVKS YLYRQWKRIL SGRVSLQDFI FAKEVRLGTY STRDSSLLPP AAIVATKSMK1680
1681 ADPRTEPRYA ERVPYVVIHG EPGARLVDMV VDPLVLLDVD TPYRLNDLYY INKQIIPALQ1740
1741 RVFGLVGADL NQWFLEMPRL TRSSLGQRPL NSKNSHKTRI DYFYLSKHCI LCGEVVQESA1800
1801 QLCNRCLQNK SAAAATIVWK TSKLEREMQH LATICRHCGG GDWVVQSGVK CNSLACSVFY1860
1861 ERRKVQKELR GLSSIATESE LYPKCMAEWF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
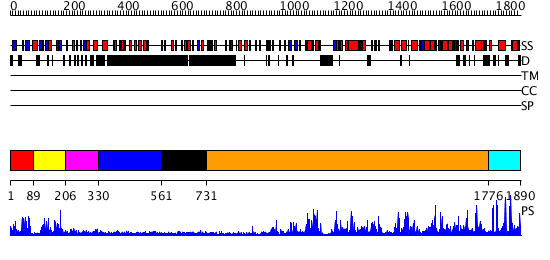
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | 5.221849 | No description for 2jm1A was found. | |
| 2 | View Details | [89..205] | 1.066978 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [206..329] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [330..560] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [561..730] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [731..1775] | 152.0 | No description for 2gv9A was found. | |
| 7 | View Details | [1776..1890] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)