













| General Information: |
|
| Name(s) found: |
AP1G1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 876 amino acids |
Gene Ontology: |
|
| Cellular Component: |
AP-1 adaptor complex
[ISS]
|
| Biological Process: |
vesicle-mediated transport
[ISS]
|
| Molecular Function: |
protein binding
[IEA]
protein transporter activity [IEA] clathrin binding [ISS] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNPFSSGTRL RDMIRAIRAC KTAAEERAVV RKECADIRAL INEDDPHDRH RNLAKLMFIH 60
61 MLGYPTHFGQ MECLKLIASP GFPEKRIGYL GLMLLLDERQ EVLMLVTNSL KQDLNHSNQY 120
121 VVGLALCALG NICSAEMARD LAPEVERLIQ FRDPNIRKKA ALCSTRIIRK VPDLAENFVN 180
181 AAASLLKEKH HGVLITGVQL CYELCTINDE ALEYFRTKCT EGLIKTLRDI TNSAYQPEYD 240
241 VAGITDPFLH IRLLRLLRVL GQGDADASDL MTDILAQVAT KTESNKNAGN AVLYECVETI 300
301 MAIEDTNSLR VLAINILGRF LSNRDNNIRY VALNMLMKAI TFDDQAVQRH RVTILECVKD 360
361 PDASIRKRAL ELVTLLVNEN NVTQLTKELI DYLEISDEDF KEDLSAKICF IVEKFSPEKL 420
421 WYIDQMLKVL CEAGKFVKDD VWHALIVVIS NASELHGYTV RALYKSVLTY SEQETLVRVA 480
481 VWCIGEYGDL LVNNVGMLGI EDPITVTESD AVDVIEDAIT RHNSDSTTKA MALVALLKLS 540
541 SRFPSISERI KDIIVKQKGS LLLEMQQRAI EYNSIVDRHK NIRSSLVDRM PVLDEATFNV 600
601 RRAGSFPASV STMAKPSVSL QNGVEKLPVA PLVDLLDLDS DDIMAAPSPS GTDFLQDLLG 660
661 VDLGSSSAQY GATQAPKAGT DLLLDILSIG TPSPAQNSTS SIGLLSIADV NNNPSIALDT 720
721 LSSPAPPHVA TTSSTGMFDL LDGLSPSPSK EATNGPAYAP IVAYESSSLK IEFTFSKTPG 780
781 NLQTTNVQAT FTNLSPNTFT DFIFQAAVPK FLQLHLDPAS SNTLLASGSG AITQNLRVTN 840
841 SQQGKKSLVM RMRIGYKLNG KDVLEEGQVS NFPRGL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
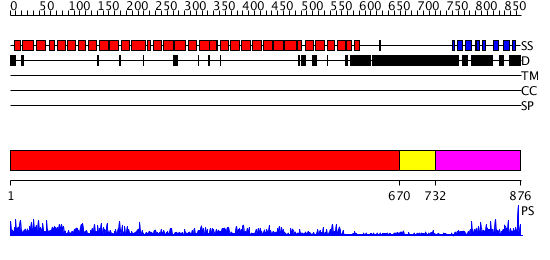
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..669] | 115.0 | AP1 clathrin adaptor core | |
| 2 | View Details | [670..731] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [732..876] | 25.0 | Gamma1-adaptin domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)