













| General Information: |
|
| Name(s) found: |
LUMI_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 953 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
vegetative to reproductive phase transition of meristem
[IMP]
positive regulation of flower development [IMP] |
| Molecular Function: |
transcription factor activity
[TAS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDAFKEEIEI GSSVESLMEL LDSQKVLFHS QIDQLQDVVV AQCKLTGVNP LAQEMAAGAL 60
61 SIKIGKRPRD LLNPKAVKYL QAVFAIKDAI SKRESREISA LFGITVAQVR EFFVTQKTRV 120
121 RKQVRLSREK VVMSNTHALQ DDGVPENNNA TNHVEPVPLN SIHPEACSIS WGEGETVALI 180
181 PPEDIPPDIS DSDKYFVENI FSLLRKEETF SGQVKLMEWI MQIQDASVLI WFLSKGGVLI 240
241 LTTWLSQAAS EEQTSVLLLI LKVLCHLPLH KASPENMSAI LQSVNGLRFY RISDISNRAK 300
301 GLLSRWTKLF AKIQAMKKQN RNSSQIDSQS QLLLKQSIAE IMGDSSNPED ILSLSNGKSE 360
361 NVRRIESSQG PKLLLTSADD STKKHMLGSN PSYNKERRKV QMVEQPGQKA AGKSPQTVRI 420
421 GTSGRSRPMS ADDIQKAKMR ALYMQSKNSK KDPLPSAIGD SKIVAPEKPL ALHSAKDSPP 480
481 IQNNEAKTED TPVLSTVQPV NGFSTIQPVN GPSAVQPVNG PLAVQPVNGP SALQPVNGPS 540
541 AVIVPVQADE IKKPSTPPKS ISSKVGVMMK MSSQTILKNC KRKQIDWHVP PGMELDELWR 600
601 VAAGGNSKEA DVQRNRNRRE RETTYQSLQT IPLNPKEPWD REMDYDDSLT PEIPSQQPPE 660
661 ESLTEPQDSL DERRIAAGAA TTSSSLSSPE PDLELLAALL KNPDLVYALT SGKPSNLAGQ 720
721 DMVKLLDVIK TGAPNSSSSS NKQVEERVEV SLPSPTPSTN PGMSGWGQEG IRNPFSRQNQ 780
781 VGTAVARSGT QLRVGSMQWH QTNEQSIPRH APSAYSNSIT LAHTEREQQQ YMQPKLHHNL 840
841 HFQQQQQQPI STTSYAVREP VGQMGTGTSS SWRSQQSQNS YYSHQENEIA SASQVTSYQG 900
901 NSQYMSSNPG YESWSPDNSP SRNQLNMRGQ QQQASRKHDS STHPYWNQNK RWR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
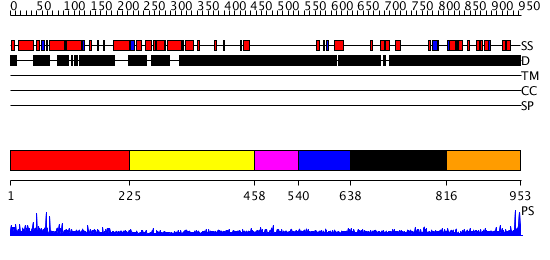
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..224] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [225..457] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [458..539] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [540..637] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [638..815] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [816..953] | 1.19899 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)