













| General Information: |
|
| Name(s) found: |
E2FC_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 396 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
nucleolus [IDA] |
| Biological Process: |
cell division
[IMP]
negative regulation of cell division [IMP] cell morphogenesis [IMP] DNA endoreduplication [IMP] |
| Molecular Function: |
DNA binding
[IDA]
transcription factor activity [ISS] protein heterodimerization activity [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAATSNSGED PTLSYHHRSP FRFELLQSIS SSDPRYSSLT PSSTNRPFSV SQSLPNSQLS 60
61 PLISPHWDDS YSQITQKVQK SRKNHRIQLG SIANMSGGES IDIAKVIVKQ ESSPQNVKRV 120
121 YNKSKGGTKL LKAGKRMANG EVQNGGLNGA SINCRYDSSL GLLTKKFVKL IQEAEDGTLD 180
181 LNYCAVVLEV QKRRIYDITN VLEGIGLIEK TTKNHIRWKG ADNLGQKDLG DQISRLKSEV 240
241 ESMQSEESRL DDLIRERQEA LRSLEEDDYC RRYMFMTEED ITSLPRFQNQ TLLAIKAPTA 300
301 SYIEVPDPDE MSFPQQYRMV IRSRMGPIDV YLLSKYKGDS AETSDKLGNE SDQKAPVGVD 360
361 TPSLKIVTSD TDLKADYWFE SDAEVSLTDL WSNFNS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
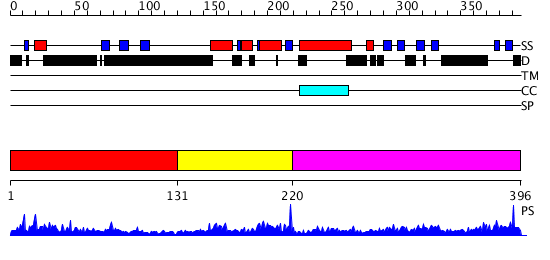
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..130] | 4.094995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [131..219] | 24.0 | Cell cycle transcription factor E2F-4 | |
| 3 | View Details | [220..396] | 21.39794 | Structure of the Rb C-terminal domain bound to an E2F1-DP1 heterodimer |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)