













| General Information: |
|
| Name(s) found: |
ECH2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 309 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA]
|
| Biological Process: |
metabolic process
[ISS]
fatty acid beta-oxidation, unsaturated, even number [IMP] |
| Molecular Function: |
oxidoreductase activity
[ISS]
3R-hydroxyacyl-CoA dehydratase activity [IDA][IGI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATSDSEFNS DLLLAHKLPE TRYTYNERDV AIYALGIGAC GQDAVDSDEL KFVYHRNGQD 60
61 LIQVLPTFAS LFTLGSLTEG LDLPGFKYDP SLLLHGQQYI EIYRPLPSKA SLINKVSLAG 120
121 LQDKGKAAIL ELETRSYEEG SGELLCMNRT TVFLRGAGGF SNSSQPFSYK NYPSNQGLAV 180
181 KIPQRQPLTV CEERTQPSQA LLYRLSGDYN PLHSDPEFAK LAGFPRPILH GLCTLGFAIK 240
241 AIIKCVCKGD PTAVKTISGR FLTTVFPGET LITEMWLEGL RVIYQTKVKE RNKTVLAGYV 300
301 DIRGLSSSL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
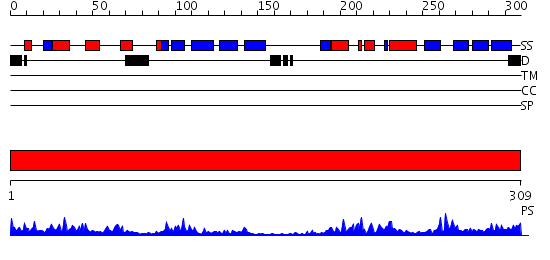
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..309] | 50.221849 | Crystal structure analysis of the 2-enoyl-CoA hydratase 2 domain of human peroxisomal multifunctional enzyme type 2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)