













| General Information: |
|
| Name(s) found: |
NUD19_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 438 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
metal ion binding
[IEA]
hydrolase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLALFLSSSS YPTLSFLSRS VTLNLARTTT LSALTMSMNL KTHAFAGNPL KSKTPKSTDP 60
61 FSPTSAFESL KTLIPVIPNH STPSPDFKVL PFSKGRPLVF SSGGDANTTP IWHLGWVSLA 120
121 DCKVLLASCG VDLNEDSLVY LGPKLEEDLV YWAVDLAEDG FVSELGGRKL CFVELRTLMV 180
181 AADWADQRAM DELAIAGNAR ALLEWHNVSQ FCGSCGSKTF PKEAGRRKQC SDETCRKRVY 240
241 PRVDPVVIML VIDRENDRAL LSRQSRYVPR MWSCLAGFIE PGESLEEAVR RETWEETGIE 300
301 VGDVVYHSSQ PWPVGPSSMP CQLMLGFFAF AKTLDINVDK EELEDAQWHS REEVKKALAV 360
361 AEYRKAQRTA AAKVEQICKG VERSQSLSTD FNLESGELAP MFIPGPFAIA HHLISAWVNQ 420
421 APDDVHSKQQ AGVSLSSL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
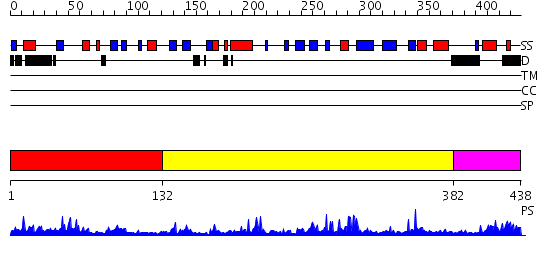
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | 7.221849 | Catalytic domain of MutY | |
| 2 | View Details | [132..381] | 39.0 | Crystal structure of NADH pyrophosphatase (1790429) from Escherichia coli k12 at 2.20 A resolution | |
| 3 | View Details | [382..438] | 25.154902 | Crystal structure of Hypothetical protein BT0354 |
| Source: Reynolds et al. 2008. Manuscript submitted |

|