













| General Information: |
|
| Name(s) found: |
gi|79383745
[NCBI NR]
gi|47550675 [NCBI NR] gi|110741647 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 855 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
protein modification process
[IEA][ISS]
|
| Molecular Function: |
protein binding
[ISS]
tubulin-tyrosine ligase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKIESFDDF VKVHGILLAA SGLPPKLYPR LFEKLAYDTF DGGDYFQIES CEDDQRRKLL 60
61 LTAESMPKDS DVFLVDHAWT FRLPDAYKQL KEIPGLAERM ASLMSVDIDV DAGEEEVAEE 120
121 LSVDQIIDNE IRYAADKGYD SLRWLELEGL GVDDSLLSLH LPSKFQDLVA LSLIGNKIES 180
181 ADVVIQEIVK FKNLKALWLN DNPVLQKSER QMADEILQGC PSLEIYNSCF TPNYGLWALG 240
241 FCGDIFGKDN PGCVQQDQPL CNVTSLDLSN RSIHNLVNKA FSVHEMPLLS HLNIRGNSLD 300
301 QNSVGELLEV LKLFPSLSSL EVDIPGPLGI NALEILESLS NLSLLNGVDT AKILETGKHV 360
361 VDSMLQPRIP ELNPDDTLVD RVLDAMWLYA LNYRLADDEK LDETSLWYVM DELGSALRHS 420
421 DEPNFKVAPF LFMPSGKLES AVSYSVMWPI KHSQKGDECT RDFLSGIGED KQRSARLTAW 480
481 FQTPENYFIH EFEKYQQKLQ AKAFESKPSN PSVSRSIQHS DGSPLLVYTD LPQVEEFLTR 540
541 PEFVITNEPK DADILWTSVQ VDDELKKEVG ITDDQYINQF PFEACLVMKH HLAETIQMGY 600
601 GSPKWLQPTY NLETQLSQFI GDYCVRKRDQ LNNLWILKPW NMARTIDTSI TDNLSAIIRM 660
661 METGPKICQK YIEHPALFKG NKFDLRYVVL VRSIDPLEIY LIEIFWVRLS NNPYSLEKHS 720
721 FFEYETHFTV MNYGRKLNHK PTAEFVREFE QEHNVKWMDI HEKVKQVIRA VFEAAALAHP 780
781 EMQSPKSRAM YGVDVMLDSS FEPKILEVTY CPDCMRACKY DMETIDGKGI VKGGDFFNNV 840
841 FGCLFLDETA HVTPL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
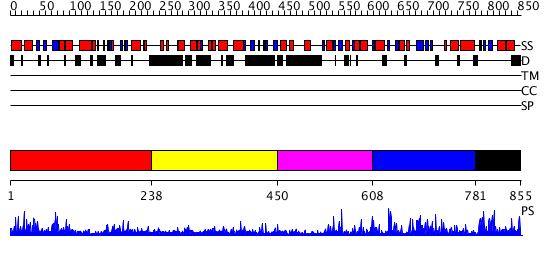
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..237] | 1.010971 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [238..449] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [450..607] | 6.264994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [608..780] | 3.28399 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [781..855] | 8.276996 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)