













| General Information: |
|
| Name(s) found: |
MARK1
[HGNC (HUGO)]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 795 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSARTPLPTV NERDTVNHTT VDGYTEPHIQ PTKSSSRQNI PRCRNSITSA TDEQPHIGNY 60
61 RLQKTIGKGN FAKVKLARHV LTGREVAVKI IDKTQLNPTS LQKLFREVRI MKILNHPNIV 120
121 KLFEVIETEK TLYLVMEYAS GGEVFDYLVA HGRMKEKEAR AKFRQIVSAV QYCHQKYIVH 180
181 RDLKAENLLL DGDMNIKIAD FGFSNEFTVG NKLDTFCGSP PYAAPELFQG KKYDGPEVDV 240
241 WSLGVILYTL VSGSLPFDGQ NLKELRERVL RGKYRIPFYM STDCENLLKK LLVLNPIKRG 300
301 SLEQIMKDRW MNVGHEEEEL KPYTEPDPDF NDTKRIDIMV TMGFARDEIN DALINQKYDE 360
361 VMATYILLGR KPPEFEGGES LSSGNLCQRS RPSSDLNNST LQSPAHLKVQ RSISANQKQR 420
421 RFSDHAGPSI PPAVSYTKRP QANSVESEQK EEWDKDVARK LGSTTVGSKS EMTASPLVGP 480
481 ERKKSSTIPS NNVYSGGSMA RRNTYVCERT TDRYVALQNG KDSSLTEMSV SSISSAGSSV 540
541 ASAVPSARPR HQKSMSTSGH PIKVTLPTIK DGSEAYRPGT TQRVPAASPS AHSISTATPD 600
601 RTRFPRGSSS RSTFHGEQLR ERRSVAYNGP PASPSHETGA FAHARRGTST GIISKITSKF 660
661 VRRDPSEGEA SGRTDTSRST SGEPKERDKE EGKDSKPRSL RFTWSMKTTS SMDPNDMMRE 720
721 IRKVLDANNC DYEQKERFLL FCVHGDARQD SLVQWEMEVC KLPRLSLNGV RFKRISGTSI 780
781 AFKNIASKIA NELKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
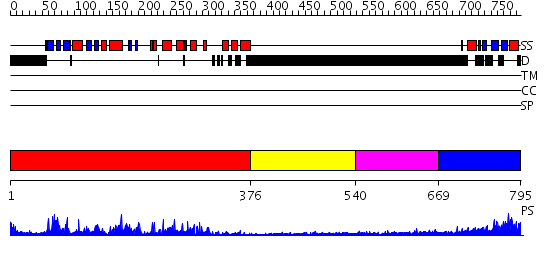
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..375] | 89.39794 | No description for 2hakA was found. | |
| 2 | View Details | [376..539] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [540..668] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [669..795] | 42.221849 | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)