













| General Information: |
|
| Name(s) found: |
SPK1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1830 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA]
plasma membrane [IDA] |
| Biological Process: | NONE FOUND |
| Molecular Function: |
GTP binding
[IEA]
GTPase binding [IEA] guanyl-nucleotide exchange factor activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MENNNLGLRF RKLPRQPLAL PKLDPLLDEN LEQWPHLNQL VQCYGTEWVK DVNKYGHYEN 60
61 IRPDSFQTQI FEGPDTDTET EIRLASARSA TIEEDVASIS GRPFSDPGSS KHFGQPPLPA 120
121 YEPAFDWENE RAMIFGQRTP ESPAASYSSG LKISVRVLSL AFQSGLVEPF FGSIALYNQE 180
181 RKEKLSEDFY FQIQPTEMQD AKLSSENRGV FYLDAPSASV CLLIQLEKTA TEEGGVTSSV 240
241 YSRKEPVHLT EREKQKLQVW SRIMPYRESF AWAVVPLFDN NLTTNTGESA SPSSPLAPSM 300
301 TASSSHDGVY EPIAKITSDG KQGYSGGSSV VVEISNLNKV KESYSEESIQ DPKRKVHKPV 360
361 KGVLRLEIEK HRNGHGDFED LSENGSIIND SLDPTDRLSD LTLMKCPSSS SGGPRNGCSK 420
421 WNSEDAKDVS RNLTSSCGTP DLNCYHAFDF CSTTRNEPFL HLFHCLYVYP VAVTLSRKRN 480
481 PFIRVELRKD DTDIRKQPLE AIYPREPGVS LQKWVHTQVA VGARAASYHD EIKVSLPATW 540
541 TPSHHLLFTF FHVDLQTKLE APRPVVVGYA SLPLSTYIHS RSDISLPVMR ELVPHYLQES 600
601 TKERLDYLED GKNIFKLRLR LCSSLYPTNE RVRDFCLEYD RHTLQTRPPW GSELLQAINS 660
661 LKHVDSTALL QFLYPILNML LHLIGNGGET LQVAAFRAMV DILTRVQQVS FDDADRNRFL 720
721 VTYVDYSFDD FGGNQPPVYP GLATVWGSLA RSKAKGYRVG PVYDDVLSMA WFFLELIVKS 780
781 MALEQARLYD HNLPTGEDVP PMQLKESVFR CIMQLFDCLL TEVHERCKKG LSLAKRLNSS 840
841 LAFFCYDLLY IIEPCQVYEL VSLYMDKFSG VCQSVLHECK LTFLQIISDH DLFVEMPGRD 900
901 PSDRNYLSSI LIQELFLSLD HDELPLRAKG ARILVILLCK HEFDARYQKA EDKLYIAQLY 960
961 FPFVGQILDE MPVFYNLNAT EKREVLIGVL QIVRNLDDTS LVKAWQQSIA RTRLYFKLME1020
1021 ECLILFEHKK AADSILGGNN SRGPVSEGAG SPKYSERLSP AINNYLSEAS RQEVRLEGTP1080
1081 DNGYLWQRVN SQLASPSQPY SLREALAQAQ SSRIGASAQA LRESLHPILR QKLELWEENV1140
1141 SATVSLQVLE ITENFSSMAA SHNIATDYGK LDCITTILTS FFSRNQSLAF WKAFFPIFNR1200
1201 IFDLHGATLM ARENDRFLKQ IAFHLLRLAV YRNDSVRKRA VIGLQILVKS SLYFMQTARL1260
1261 RALLTITLSE LMSDVQVTHM KSDNTLEESG EARRLQQSLS EMADEAKSVN LLRECGLPDD1320
1321 TLLIIPEKFT ENRWSWAEVK HLSDSLVLAL DASLGHALLG SVMAMDRYAA AESFYKLGMA1380
1381 FAPVPDLHIM WLLHLCDAHQ EMQSWAEAAQ CAVAVAGVIM QALVARNDGV WSKDHVSALR1440
1441 KICPMVSGEF TTEASAAEVE GYGASKLTVD SAVKYLQLAN KLFSQAELYH FCASILELVI1500
1501 PVYKSRKAYG QLAKCHTLLT NIYESILDQE SNPIPFIDAT YYRVGFYGEK FGKLDRKEYV1560
1561 YREPRDVRLG DIMEKLSHIY ESRMDSNHIL HIIPDSRQVK AEDLQAGVCY LQITAVDAVM1620
1621 EDEDLGSRRE RIFSLSTGSV RARVFDRFLF DTPFTKNGKT QGGLEDQWKR RTVLQTEGSF1680
1681 PALVNRLLVT KSESLEFSPV ENAIGMIETR TTALRNELEE PRSSDGDHLP RLQSLQRILQ1740
1741 GSVAVQVNSG VLSVCTAFLS GEPATRLRSQ ELQQLIAALL EFMAVCKRAI RVHFRLIGEE1800
1801 DQEFHTQLVN GFQSLTAELS HYIPAILSEL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
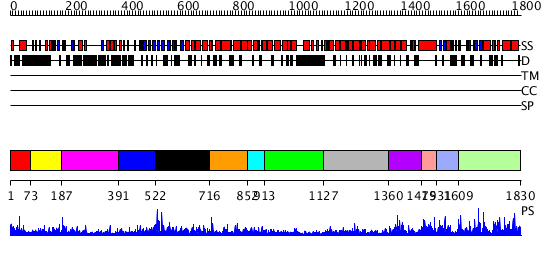
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..72] | 12.522879 | Solution structure of pleckstrin homology domain from human KIAA1058 protein | |
| 2 | View Details | [73..186] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [187..390] | 3.111994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [391..521] | 1.123995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [522..715] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [716..851] | 1.143996 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [852..912] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [913..1126] | 1.161996 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1127..1359] | 1.164979 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1360..1478] | 1.170987 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1479..1530] | 2.169992 | View MSA. No confident structure predictions are available. | |
| 12 | View Details | [1531..1608] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [1609..1830] | 114.619789 | No description for PF06920.5 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)