













| General Information: |
|
| Name(s) found: |
HEM2_ORYSJ
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 426 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASTVSFSPA NVQMLQGRSC HGHAAFGGCS AVPRTGPRMR SVAVRVSSEQ EAAPAVRAPS 60
61 GRTIEECEAD AVAGRFPAPP PLVRPKAPEG TPQIRPLDLT KRPRRNRRSP ALRAAFQETT 120
121 ISPANLVLPL FIHEGEDDAP IGAMPGCYRL GWRHGLLDEV YKSRDVGVNS FVLFPKVPDA 180
181 LKSQSGDEAY NDNGLVPRTI RLLKDKFPDI VVYTDVALDP YSSDGHDGIV REDGVIMNDE 240
241 TVYQLCKQAV SQARAGADVV SPSDMMDGRV GAIRAALDAE GFHDVSIMSY TAKYASSFYG 300
301 PFREALDSNP RFGDKKTYQM NPANYREALL ETAADEAEGA DILLVKPGLP YLDVIRLLRD 360
361 NSALPIAAYQ VSGEYSMIKA GGALNMIDEE KVMMESLMCL RRAGADIILT YFARQAANVL 420
421 CGMRSN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
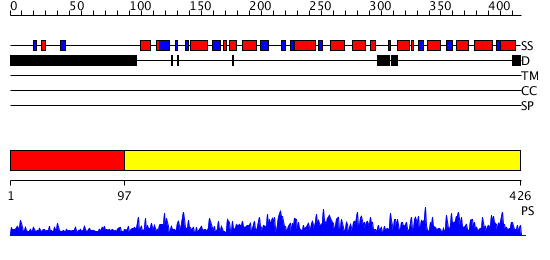
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [97..426] | 149.0 | Structure of the plant like 5-Aminolaevulinic Acid Dehydratase from Chlorobium vibrioforme |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)