













| General Information: |
|
| Name(s) found: |
ARFR_ORYSJ
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 700 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MITFVDSAAK ERERESDKCL DPQLWHACAG GMVQMPPVSS KVYYFPQGHA EHAQGHGPVE 60
61 FPGGRVPALV LCRVAGVRFM ADPDTDEVFA KIRLVPVRAN EQGYAGDADD GIGAAAAAAA 120
121 QEEKPASFAK TLTQSDANNG GGFSVPRYCA ETIFPRLDYS ADPPVQTVLA KDVHGVVWKF 180
181 RHIYRGTPRR HLLTTGWSTF VNQKKLVAGD SIVFMRTENG DLCVGIRRAK KGGVGGPEFL 240
241 PPPPPPPPTP AAGGNYGGFS MFLRGDDDGN KMAAAARGKV RARVRPEEVV EAANLAVSGQ 300
301 PFEVVYYPRA STPEFCVKAG AVRAAMRTQW FAGMRFKMAF ETEDSSRISW FMGTVSAVQV 360
361 ADPIRWPNSP WRLLQVSWDE PDLLQNVKRV SPWLVELVSN MPAIHLAPFS PPRKKLCVPL 420
421 YPELPIDGQF PTPMFHGNPL ARGVGPMCYF PDGTPAGIQG ARHAQFGISL SDLHLNKLQS 480
481 SLSPHGLHQL DHGMQPRIAA GLIIGHPAAR DDISCLLTIG SPQNNKKSDG KKAPAQLMLF 540
541 GKPILTEQQI SLGDAASVDV KKSSSDGNAE NTVNKSNSDV SSPRSNQNGT TDNLSCGGVP 600
601 LCQDNKVLDV GLETGHCKVF MQSEDVGRTL DLSVVGSYEE LYRRLADMFG IEKAELMSHV 660
661 FYRDAAGALK HTGDEPFSEF TKTARRLNIL TDTSGDNLAR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
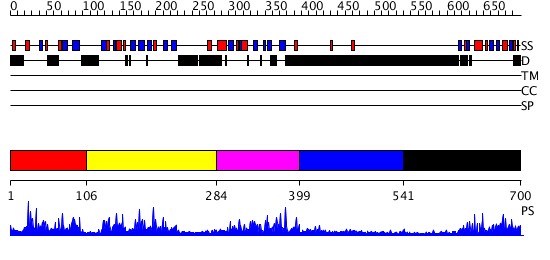
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | 2.070991 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [106..283] | 39.522879 | Solution Structure of the B3 DNA-Binding Domain of RAV1 | |
| 3 | View Details | [284..398] | 2.113983 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [399..540] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [541..700] | 1.12 | Solution structure of RSGI RUH-024, a PB1 domain in human cDNA, KIAA0049 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)