













| General Information: |
|
| Name(s) found: |
gi|50251645
[NCBI NR]
gi|222641216 [NCBI NR] gi|115478352 [NCBI NR] gi|113631004 [NCBI NR] |
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 1740 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEGELGQQT VELGAVVRRA AEESYLSLRE LVEKSQDEGE GKGGAYGARQ RSDSEKKIDL 60
61 LKFIARTRQR MLRLHVLAKW CQQVPLVNYC QQLGSTLSSH ETCFTQTADS LYFMHEGLQQ 120
121 ARAPMFDIPS ALEVMLTGNY QRLPLCIEDI GSQNKLSPDE EKRALQKLNA SVRYKVLVTP 180
181 RPKEVSNVSV ADGIAVFRVD GEFKVLLTLG YRGHLDLWRI LHLEVLVGDK GGPIKLEERR 240
241 RFALGDDIER RMAVSENPFM VLYAILHELC ISLAMDTIIR KTNVLRQGRW KDAIRSELVL 300
301 DSTTGQTGSA PLMQLGQDGE YDLSGSRIPG LKINYWLDEK AGGSAESDSS PFIKIEARQD 360
361 MQIKCQHSSF ILDPLTDKEA NLSLDLCCID VEQLILRAIA SNRHTRLLDI QRQLSKNVQI 420
421 SQSPKDVILK RDVEIAKDPV KKTEQKDFAD CCGNEVLQVR AYGQAYIGLG INIRSGRFLL 480
481 QSPENILPPS ALLDCEEALN KGSITATDVF ASLRTRSILH LFAATGSFFG LKVYEKSQGT 540
541 LKIPKDILHG SDLMVMGFPQ CANAYYLLMQ LDKDFRPVFH LLETQSDAND KTNTNACTKE 600
601 ALRFNKIDIG QIQISKSESN TNMFDAKLHA LQNIGNCDDV MENRLPIQSG IEPLPLLPAC 660
661 SPSFSSVVDE VFEYEHGALA VPNHSLPQTT LQSTSHPGSL SVGFQGVGTR ANASIEGASS 720
721 AYSGSKFSPG VGLNSYLPSN LRHVQSTNAF SSSTVTKSSS IKLPSSNSNH ELSSLSSPTE 780
781 HVIADGSKSL QLVPASKING SINLITMGSD GASRKRSISD LFLNLPSLQG LKPSSPRKRR 840
841 RISESMESWS PLQAYSSDSQ SRTSLTYGNI LAERNNCVPA TTYASVLLHV IRHCSLSIKH 900
901 AQITAQMDSL AIPYVEEVGL RSPSSNLWLR LPFARDDSWK HICLRLGKAG SMSWDVRIND 960
961 PHFRELWELS TGSTTTSWGV GVRVANTSEM DSHISFDAEG VILTYSNVEP DSVQKLVSDL1020
1021 RRLANARSFA RGMRRLIGVK LNDKLDDDQT STDIKSQPVN KGNSDAADRL SEQMRKTFRI1080
1081 EAVGLMSLWF SYGTMPMVHF VVEWESAKGG CTMHVSPDQL WPHTKFLEDF VNGGEVASFL1140
1141 DCIRLTAGPL LALGGAIRPA RMPVTVSSGY NSMPKQMNNI PTQGPLANGS SSTTMHHAPS1200
1201 PANVAATHLG SHNLHTAAML SAAGRGGPGL VPSSLLPFDV SVVLRGPYWI RIIYRKKFSV1260
1261 DMRCFAGDQV WLQPATPPKG GPLVGGSLPC PQFRPFIMEH VAQGLNALEP SFMNATQAGA1320
1321 HLNSSAGTLQ PAPNANRVNA TQGIGMSRPA SGVANHVAAN LSRAGNAMLA SSGLASGIGG1380
1381 ASVRLTSGAN LPVHVKGELN TAFIGLGDDG GYGGGWVPLA ALKKVLRGIL KYLGVLWLFA1440
1441 QLPELLKEIL GSILKENEGA LLNLDQEQPA LRFYVGGYVF AVSVHRVQLL LQVLSVKRFH1500
1501 HQQQQQQAQN SAQEELAPPE INEICDYFSR RVASEPYDAS RVASFITLLT LPISVLREFL1560
1561 KLIAWKKGFS QAHGDMATAQ RARIEICLEN HSGSVSDDIT ESTLAKSNVK YDRAHSSLEF1620
1621 ALTFVLDHAL IPHMNVAGGA AWLPYCVSVR LKYSFGESNH IAYLAMDGSH GGRACWLQYE1680
1681 DWERCKQKVA RAVETVNGSV AVGESGQGRL RMVAEMIQKQ LQLCLQQLRD GPLSAGSTAS1740
1741 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
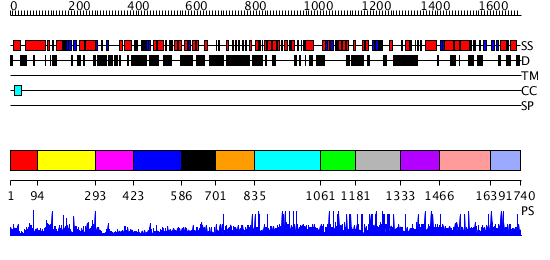
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | 1.044988 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [94..292] | 1.048974 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [293..422] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [423..585] | 1.038991 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [586..700] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [701..834] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [835..1060] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1061..1180] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1181..1332] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1333..1465] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1466..1638] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1639..1740] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)