













| General Information: |
|
| Name(s) found: |
XRN2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1012 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
miRNA catabolic process
[IMP]
|
| Molecular Function: |
5'-3' exonuclease activity
[ISS]
nucleic acid binding [ISS] 5'-3' exoribonuclease activity [IGI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVPSFYRWL IQRYPLTIQE VIEEEPLEVN GGGVTIPIDS SKPNPNGYEY DNLYLDMNGI 60
61 IHPCFHPEDK PSPTTFTEVF QCMFDYIDRL FVMVRPRKLL FMAIDGVAPR AKMNQQRARR 120
121 FRAAKDAAEA AAEEEQLREE FEREGKKLPP KVDSQVFDSN VITPGTEFMA TLSFALRYYI 180
181 HVRLNSDPGW KNIKVILSDA NVPGEGEHKI MSYIRCNKNH PGYNPNTHHC LYGLDADLIM 240
241 LSLATHEIHF SILREVVFFP GEEGKCFLCG QEGHRAADCE GKIKRKTGEM LDNTEADVVV 300
301 KKPYEFVNIW ILREYLEHDM QIPGAKKNLD RLIDDFIFIC FFVGNDFLPH MPTLEIREGA 360
361 IELLMSVYKN KFRSAKKYLT DSSKLNLRNV ERFIKAVGMY ENQIFQKRAQ VQQRQSERFR 420
421 RDKARDKARD NARDNAQASR QFSGKLVQLD SLDEVSDSLH SSPSRKYLRL SLDDNIGVAN 480
481 VETENSLKAE ELDNEEDLKF KLKKLLRDKG DGFRSGNGEQ DKVKLNKVGW RERYYEEKFA 540
541 AKSVEEMEQI RRDVVLKYTE GLCWIMHYYY HGVCSWNWFY PYHYAPFASD LKGLEKLDIK 600
601 FELGSPFKPF NQLLAVLPSA SAHALPECYR SLMTNPDSPI ADFYPADFEI DMNGKRYSWQ 660
661 GISKLPFVEE KRLLEAAAQV EKSLTNEEIR RNSALFDMLF VVASHPLGEL IRSLNSRTNN 720
721 LSNEERATII EKIDPGLSDG MNGYIASCGG DSQPSCFCST VEGMEDVLTN QVICAIYKLP 780
781 EDIRGSEITH QIPRLAIPKK TISLVDLKSG GLLWHEDGDK RRAPPKVIKI KRYNPEGSIS 840
841 GGRLGKASHR LVLQTINAQP DYMNINSEPA LCPNTVFQNE RVPKKIPTFK DNGIQWISPP 900
901 PSQITPKKMN SPQRQKAWKK DETPQSREKS KKLKSSLKVN PLKMKKTKSP QREFTREKKK 960
961 ENITPQRKLT KAQRQVKHIR MMEEAKMIKQ RKKEKYLRKK AKYAQGAPPK TA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
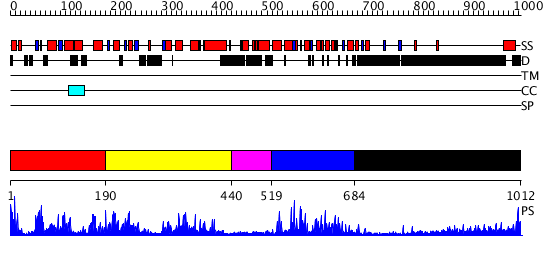
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..189] | 2.119939 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [190..439] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [440..518] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [519..683] | 2.107963 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [684..1012] | 7.39794 | No description for 1y0fB was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)