













| General Information: |
|
| Name(s) found: |
EDR2L_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 719 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IEA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
lipid binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKVVYEGWM VRYGRRKIGR SYIHMRYFVL EPRLLAYYKK KPQDNQLPIK TMVIDGNCRV 60
61 EDRGLKTHHG HMVYVLSIYN KKEKHHRITM AAFNIQEALM WKEKIECVID QHQDSLVPSG 120
121 QQYVSFEYKP GMDAGRTASS SDHESPFSAL EDENDSQRDL LRRTTIGNGP PESILDWTKE 180
181 FDAELSNQSS SNQAFSRKHW RLLQCQNGLR IFEELLEVDY LPRSCSRAMK AVGVVEATCE 240
241 EIFELVMSMD GTRYEWDCSF HNGRLVEEVD GHTAILYHRL LLDWFPMVVW PRDLCYVRYW 300
301 RRNDDGSYVV LFRSREHENC GPQPGFVRAH LESGGFNIAP LKPRNGRPRT QVQHLIQIDL 360
361 KGWGSGYLPA FQQHCLLQML NSVSGLREWF SQTDDRGQPI RIPVMVNMAS SSLALGKGGK 420
421 HHHKSSLSID QTNGASRNSV LMDEDSDDDD EFQIPDSEPE PETSKQDQET DAKKTEEPAL 480
481 NIDLSCFSGN LRHDDNENAR NCWRISDGNN FKVRGKSFCD DKRKIPAGKH LMDLVAVDWF 540
541 KDTKRMDHVV RRKGCAAQVA AEKGLFSTVV NVQVPGSTHY SMVFYFVTKE LVPGSLFQRF 600
601 VDGDDEFRNS RLKLIPLVPK GSWIVRQSVG STPCLLGKAV DCNYIRGPTY LEIDVDIGSS 660
661 TVANGVLGLV IGVITSLVVE MAFLVQANTP EELPERLIGA VRVSHVELSS AIVPNLDSD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
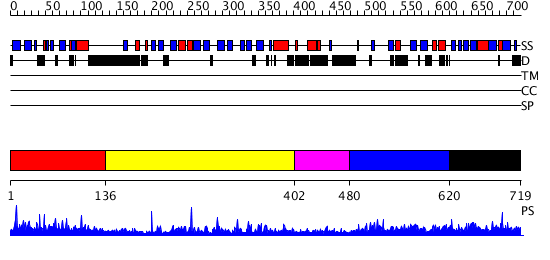
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | 5.22 | No description for 2ys3A was found. | |
| 2 | View Details | [136..401] | 33.69897 | No description for 2r55A was found. | |
| 3 | View Details | [402..479] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [480..619] | 3.050994 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [620..719] | 2.028973 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)