













| General Information: |
|
| Name(s) found: |
NUDT2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 278 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[RCA]
|
| Biological Process: |
response to oxidative stress
[IMP]
|
| Molecular Function: |
ADP-ribose diphosphatase activity
[IDA]
NAD or NADH binding [IDA] hydrolase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSASSSSTNP MSREDATTLL PSVQDKYGGV MTEMTHPMDP SLFSTLLRSS LSTWTLQGKK 60
61 GVWIKLPKQL IGLAETAVKE GFWFHHAEKD YLMLVYWIPK EDDTLPANAS HRVGIGAFVI 120
121 NHNKEVLVVQ EKTGRFQGQG IWKFPTGVVN EGEDIHDGSV REVKEETGVD TEFDQILAFR 180
181 QTHKAFFGKS DLFFVCMLKP LSLEINAQES EIEAAQWMPW EEYINQPFVQ NYELLRYMTD 240
241 ICSAKTNGDY EGFTPLRVSA PDQQGNLYYN TRDLHSRN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
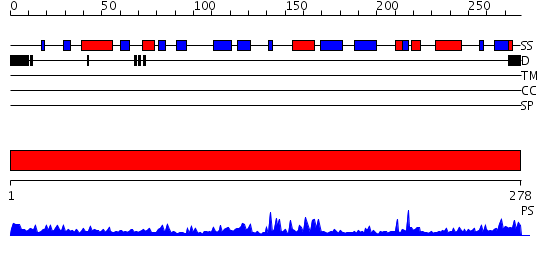
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..278] | 29.0 | Crystal structure of NADH pyrophosphatase (1790429) from Escherichia coli k12 at 2.20 A resolution |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)