













| General Information: |
|
| Name(s) found: |
QSOX1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 528 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to cation stress
[IMP]
|
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] thiol oxidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLIHLFLLL GLLSLEAAAS FSPGSRSILR DIGSNVADQK DNAIELNATN FDSVFQDSPA 60
61 KYAVLEFFAH WCPACRNYKP HYEKVARLFN GADAVYPGVV LMTRVDCAIK MNVKLCDKFS 120
121 INHYPMLFWA PPKRFVGGSW GPKQEKNEIS VVNEWRTADL LLNWINKQIG SSYGLDDQKL 180
181 GNLLSNISDQ EQISQAIFDI EEATEEAFDI ILAHKAIKSS ETSASFIRFL QLLVAHHPSR 240
241 RCRTGSAEIL VNFDDICPSG ECSYDQESGA KDSLRNFHIC GKDVPRGYYR FCRGSKNETR 300
301 GFSCGLWVLM HSLSVRIEDG ESQFAFTAIC DFINNFFMCD DCRRHFHDMC LSVKTPFKKA 360
361 RDIALWLWST HNKVNERLKK DEDSLGTGDP KFPKMIWPPK QLCPSCYLSS TEKNIDWDHD 420
421 QVYKFLKKYY GQKLVSVYKK NGESVSKEEV IAAAEEMAVP TNALVVPVGA ALAIALASCA 480
481 FGALACYWRT QQKNRKYNYN PHYLKRYNSN YMVMNTFSNT ESEREKER |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
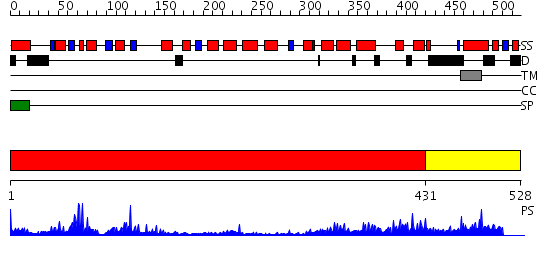
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..430] | 27.0 | Crystal Structure of Yeast Protein Disulfide Isomerase | |
| 2 | View Details | [431..528] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|