













| General Information: |
|
| Name(s) found: |
VAB_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 498 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
signal transduction
[ISS]
|
| Molecular Function: |
phosphoinositide binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERELRRPNP INSSRRPEIF SSGGSSTQLP ESPRGPMEFL SRSWSVSALE VSRALHTAKS 60
61 ASATNRPPSS INTPIPEETL NPEKEECPPE NSTSVSSQFS FAASATSQLV LERIMSQSEV 120
121 SPLTSGRLSH SSGPLNGGGS FTETDSPPIS PSDEFDDVIK YFRTHNTIHP LFSGTGGSRG 180
181 TTGNGSNTPM AGTGPKTVGR WLKDRKEKKK EETRTQNAQV HAAVSVAAVA SAVAAVAAAT 240
241 AASSPGKNEQ MARIDMAMAS AAALVAAQCV EAAEIMGADR DHLTSVVSSA VNVKSHDDIV 300
301 TLTAAAATAL RGAATLKARA LKEVWNIAAV LPAEKGASSA LCGQVDTKHS DSSFSGELPV 360
361 AGEDFLGVCN QELLAKGTEL LKRTRGGELH WKIVSVYINK AGQAVLKMKS KHVGGTFTKK 420
421 KKHMVLEVRK DIPAWAGRDL FNGDKHHYFG LKTETKRVIE FECRNQREYE IWTQGVSRLL 480
481 AIAAEKKQKS SMSKWMAP |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
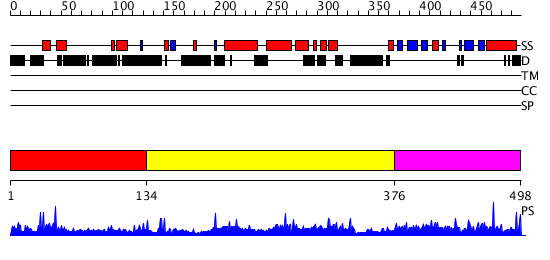
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [134..375] | 1.024983 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [376..498] | 70.0 | No description for PF08458.1 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.43 |
Source: Reynolds et al. (2008)