













| General Information: |
|
| Name(s) found: |
TAF1B_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1786 amino acids |
Gene Ontology: |
|
| Cellular Component: |
transcription factor TFIID complex
[TAS]
membrane [IDA] |
| Biological Process: |
histone acetylation
[IMP]
response to light stimulus [IEP] |
| Molecular Function: |
DNA binding
[ISS]
histone acetyltransferase activity [ISS] transcription cofactor activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MICRVDYGSN DEEYDGPELQ VVTEEDHLLP KREYLSAAFA LSGLNSRASV FDDEDYDEQG 60
61 GQEKEHVPVE KSFDSEEREP VVLKEEKPVK HEKEASILGN KNQMDTGDVQ EELVVGLSEA 120
121 TLDEKRVTPL PTLYLEDDGM VILQFSEIFA IQEPQKKRQK REIRCITYRD KYISMDISEL 180
181 IEDDEEVLLK SHGRIDTHGK KTDQIQLDVP LPIRERSQLV KSGIVRDTTS ESREFTKLGR 240
241 DSCIMGELLK QDLKDDNSSL CQSQLTMEVF PLDQQEWEHL ILWEISPQFS ANCCEGFKSG 300
301 LESAGIMVQV RASNSVTEQE SLNVMNSGGQ TQGDNNNMLE PFFVNPLESF GSRGSQSTNE 360
361 STNKSRHHPQ LLRLESQWDE DHYRENGDAG RENLKQLNSD ARGRLSGLAL QDRDMWDESW 420
421 LDSIIWESDK DLSRSKLIFD LQDEQMIFEV PNNKERKYLQ LHAGSRIVSR SSKSKDGSFQ 480
481 EGCGSNSGWQ FNISNDKFYM NGKSAQKLQG NAKKSTVHSL RVFHSAPAIK LQTMKIKLSN 540
541 KERANFHRPK ALWYPHDNEL AIKQQKILPT QGSMTIVVKS LGGKGSLLTV GREESVSSLK 600
601 AKASRKLDFK ETEAVKMFYM GKELEDEKSL AEQNVQPNSL VHLLRTKVHL WPWAQKLPGE 660
661 NKSLRPPGAF KKKSDLSNQD GHVFLMEYCE ERPLMLSNAG MGANLCTYYQ KSSPEDQHGN 720
721 LLRNQSDTLG SVIILEHGNK SPFLGEVHGG CSQSSVETNM YKAPVFPHRL QSTDYLLVRS 780
781 AKGKLSLRRI NKIVAVGQQE PRMEIMSPAS KNLHAYLVNR MMAYVYREFK HRDRIAADEL 840
841 SFSFSNISDA TVRKYMQVCS DLERDANGKA CWSKKRKFDK IPLGLNTLVA PEDVCSYESM 900
901 LAGLFRLKHL GITRFTLPAS ISTALAQLPD ERIAAASHIA RELQITPWNL SSSFVTCATQ 960
961 GRENIERLEI TGVGDPSGRG LGFSYVRVAP KSSAASEHKK KKAAACRGVP TVTGTDADPR1020
1021 RLSMEAAREV LLKFNVPDEI IAKQTQRHRT AMIRKISSEQ AASGGKVGPT TVGMFSRSQR1080
1081 MSFLQLQQQA REMCHEIWDR QRLSLSACDD DGNESENEAN SDLDSFVGDL EDLLDAEDGG1140
1141 EGEESNKSMN EKLDGVKGLK MRRWPSQVEK DEEIEDEAAE YVELCRLLMQ DENDKKKKKL1200
1201 KDVGEGIGSF PPPRSNFEPF IDKKYIATEP DASFLIVNES TVKHTKNVDK ATSKSPKDKQ1260
1261 VKEIGTPICQ MKKILKENQK VFMGKKTARA NFVCGACGQH GHMKTNKHCP KYRRNTESQP1320
1321 ESMDMKKSTG KPSSSDLSGE VWLTPIDNKK PAPKSATKIS VNEATKVGDS TSKTPGSSDV1380
1381 AAVSEIDSGT KLTSRKLKIS SKAKPKASKV ESDSPFHSLM PAYSRERGES ELHNPSVSGQ1440
1441 LLPSTETDQA ASSRYTTSVP QPSLSIDKDQ AESCRPHRVI WPPTGKEHSQ KKLVIKRLKE1500
1501 ITDHDSGSLE ETPQFESRKT KRMAELADFQ RQQRLRLSEN FLDWGPKDDR KWRKEQDIST1560
1561 ELHREGKVRR AYDDSTVSEE RSEIAESRRY REVIRSEREE EKRRKAKQKK KLQRGILENY1620
1621 PPRRNDGISS ESGQNINSLC VSDFERNRTE YAPQPKRRKK GQVGLANILE SIVDTLRVKE1680
1681 VNVSYLFLKP VTKKEAPNYL EIVKCPMDLS TIRDKVRRME YRDRQQFRHD VWQIKFNAHL1740
1741 YNDGRNLSIP PLADELLVKC DRLLDEYRDE LKEAEKGIVD SSDSLR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
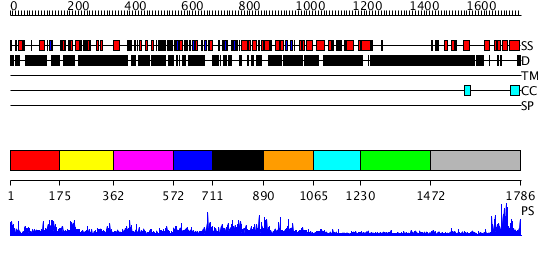
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..174] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [175..361] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [362..571] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [572..710] | 23.30103 | No description for 2ojrA was found. | |
| 5 | View Details | [711..889] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [890..1064] | 17.30103 | No description for 3dtpB was found. | |
| 7 | View Details | [1065..1229] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1230..1471] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1472..1786] | 26.0 | TAFII250 double bromodomain module |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)