













| General Information: |
|
| Name(s) found: |
gi|168278889
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | synthetic construct |
| Length: | 672 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTPSQVAFEI RGTLLPGEVF AICGSCDALG NWNPQNAVAL LPENDTGESM LWKATIVLSR 60
61 GVSVQYRYFK GYFLEPKTIG GPCQVIVHKW ETHLQPRSIT PLESEIIIDD GQFGIHNGVE 120
121 TLDSGWLTCQ TEIRLRLHYS EKPPVSITKK KLKKSRFRVK LTLEGLEEDD DDRVSPTVLH 180
181 KMSNSLEISL ISDNEFKCRH SQPECGYGLQ PDRWTEYSIQ TMEPDNLELI FDFFEEDLSE 240
241 HVVQGDALPG HVGTACLLSS TIAESGKSAG ILTLPIMSRN SRKTIGKVRV DYIIIKPLPG 300
301 YSCDMKSSFS KYWKPRIPLD VGHRGAGNST TTAQLAKVQE NTIASLRNAA SHGAAFVEFD 360
361 VHLSKDFVPV VYHDLTCCLT MKKKFDADPV ELFEIPVKEL TFDQLQLLKL THVTALKSKD 420
421 RKESVVQEEN SFSENQPFPS LKMVLESLPE DVGFNIEIKW ICQQRDGMWD GNLSTYFDMN 480
481 LFLDIILKTV LENSGKRRIV FSSFDADICT MVRQKQNKYP ILFLTQGKSE IYPELMDLRS 540
541 RTTPIAMSFA QFENLLGINV HTEDLLRNPS YIQEAKAKGL VIFCWGDDTN DPENRRKLKE 600
601 LGVNGLIYDR IYDWMPEQPN IFQVEQLERL KQELPELKSC LCPTVSRFVP SSLCGESDIH 660
661 VDANGIDNVE NA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
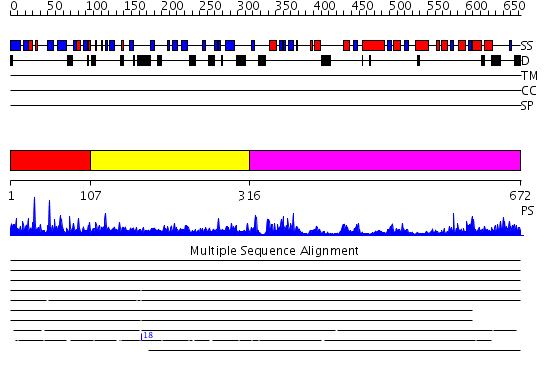
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 30.69897 | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN COMPLEX WITH CYCLODEXTRIN, NMR, MINIMIZED AVERAGE STRUCTURE | |
| 2 | View Details | [107..315] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [316..672] | 54.221849 | Crystal structure of periplasmic glycerophosphodiester phosphodiesterase from Escherichia coli |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)