













| General Information: |
|
| Name(s) found: |
gi|190406177
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 918 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIKTRIEEVQ LQFLTGNTEL THLKVSNDQL IVTTQRTIYR INLQDPAIVN HFDCPLSKEL 60
61 ETIMNVHVSP MGSVILIRTN FGRYMLLKDG EFTQLNKIKN LDLSSLHWIN ETTFLMGIKK 120
121 TPKLYRVELT GKDITTKLWY ENKKLSGGID GIAYWEGSLL LTIKDNILYW RDVTNMKFPL 180
181 VLPDESEQFE RLKHHAIKKF DSYNGLFAWV TSNGIVFGDL KEKQMEKDPA SNNFGKFLSS 240
241 SKVLLNFELP DYQNDKDHLI KDIVLTAFHI LLLRKNTVTM VSQLNNDVVF HETIPRHQLT 300
301 GSNTDSNEKF LGLVRDSVKE TFWCFSNINV FEIIIENEPN SVWNLLVRDN KFDKALSLKG 360
361 LTVREIESVK LSKAMYLFHT AKDFHSAAQT LGSMKDLSHF GEIALNFLQI KDYNDLNVIL 420
421 IKQLDNVPWK STQVVLSSWI IWNFMKQLND IELKINTTKP ASTDEDNLLN WNLNLKEKSN 480
481 ELTKFLESHL EKLDNETVYQ IMSKQNRQNE LLIFASLIND MKFLLSFWID QGNWYESLKI 540
541 LLTINNHDLV YKYSLILLLN SPEATVSTWM KIKDLDPNKL IPTILKFFTN WQNNSKLITN 600
601 ISEYPENYSL TYLKWCVREV PKMCNPIVYN SILYMMITDP RNDMILENDI IKFMKSNENK 660
661 YDLNFQLRLS LKFKKTKTSI FLLTRLNLFE DAIDLALKNN LIDDCKVIVN DEILIEDYKL 720
721 RKRLWLKIAK HLLLSMKDID IKQLIRTILN DSNEILTIKD LLPFFNEYTT IANLKEELIK 780
781 FLENHNMKMN EISEDIINSK NLKVEINTEI SKFNEIYRIL EPGKSCDECG KFLQIKKFIV 840
841 FPCGHCFHWN CIIRVILNSN DYNLRQKTEN FLKAKSKHNL NDLENIIVEK CGLCSDININ 900
901 KIDQPISIDE TELAKWNE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
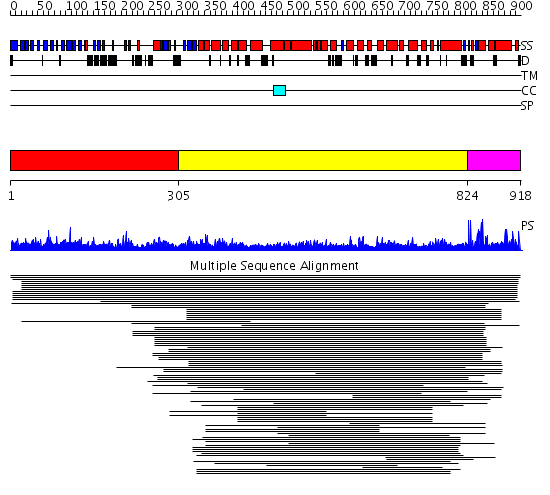
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..304] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [305..823] | 21.30103 | Clathrin heavy chain proximal leg segment | |
| 3 | View Details | [824..918] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [816..918] | 1.57 | No description for 2ecnA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)