













| General Information: |
|
| Name(s) found: |
gi|190408309
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 741 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVDTAPYIGS LGRSSLFDTG DIEQAPGNNA IGINEEDIHA FVSSTGETVQ LKKKPAKLAT 60
61 GNISLYTNPD TVWRSDDTYG ININYLLDKI EASGDDRTNA QKTSPITGKI GSDTLWVEKW 120
121 RPKKFLDLVG NEKTNRRMLG WLRQWTPAVF KEQLPKLPTE KEVSDMELDP LKRPPKKILL 180
181 LHGPPGIGKT SVAHVIAKQS GFSVSEINAS DERAGPMVKE KIYNLLFNHT FDTNPVCLVA 240
241 DEIDGSIESG FIRILVDIMQ SDIKATNKLL YGQPDKKDKK RKKKRSKLLT RPIICICNNL 300
301 YAPSLEKLKP FCEIIAVKRP SDTTLLERLN LICHKENMNI PIKAINDLID LAQGDVRNCI 360
361 NNLQFLASNV DSRDSSASDK PACAKNTWAS SNKDSPISWF KIVNQLFRKD PHRDIKEQFY 420
421 ELLNQVELNG NSDRILQGCF NIFPYVKYSD NGIRKPANIS DWLFFHDLMY QSMYAHNGEL 480
481 LRYSALVPLV FFQTFGDIAN KDDIRMKNSE YEQRELKRAN SDIVSLIMRH ISVQSPLMAS 540
541 FTDRKSLIFE ILPYLDSMIS SDFNKIRNLK LKQAIMEELV QLLKSFQLNL IQNRSEGFDV 600
601 RGGLTIDPPI DEVVLLNPKH INEVQHKRAN NLSSLLAKIE ENRAKKRHID QVTEDRLQSQ 660
661 EMHSKKVKTG LNSSSSTIDF FKNQYGLLKQ TQELEETQKT IGSDETNQAD DCNQTVKIWV 720
721 KYNEGFSNAV RKNVTWNNLW E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
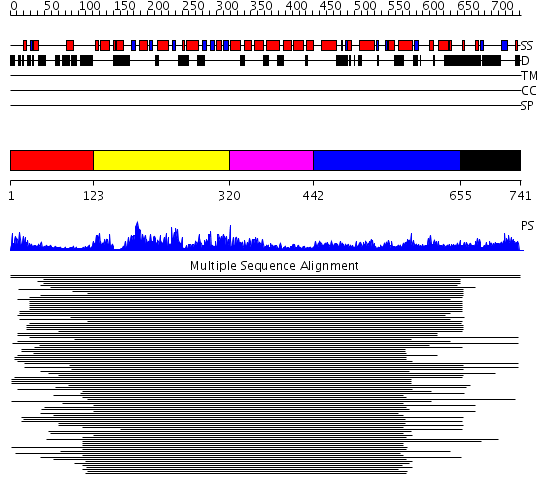
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | 104.165202 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 2 | View Details | [123..319] | 104.165202 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 3 | View Details | [320..441] | 104.165202 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 4 | View Details | [442..654] | 1.333991 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [655..741] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)