













| General Information: |
|
| Name(s) found: |
gi|190406880
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 950 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFFDSLRQK APFLDKLADS FTPTLTRDEK FRLKYKLPAN ENILEDTNAE VSFATSIKDG 60
61 KGHSDRVNNK GRKTAYVYSG RLFLTPHFLV FRDAFDHSSC VLILNISTIK RVERSPSESY 120
121 EFALLVTLYT GAKVLIQFIG IRYRSEQFCD KLKLNLKENI PNAKTLPAFL ETSYSEFLIA 180
181 KNILGKKDIT VPRAGLGQHF KYPGNPTMVK EKAKLRLWFD YFRENGRNLA VVQTPMFRKL 240
241 IRIGVPNRMR GEIWELCSGA MYMRYANSGE YERILNENAG KTSQAIDEIE KDLKRSLPEY 300
301 SAYQTEEGIQ RLRNVLTAYS WKNPDVGYCQ AMNIVVAGFL IFMSEEQAFW CLCNLCDIYV 360
361 PGYYSKTMYG TLLDQRVFES FVEDRMPVLW EYILQHDIQL SVVSLPWFLS LFFTSMPLEY 420
421 AVRIMDIFFM NGSITLFQVA LAVLKINADD ILQADDDGMF IAIIKHYFQT LGQSAHPDSS 480
481 DIKYRQITKF QELLVTAFKE FSVISEEMAM HARHKYEKGI FQNIETFMKR TQLRHMPKTF 540
541 NLSSDDLSNI YDMFYQSIET YKISMGTGSS NMGFEVFIQF LSKFCDSCRP CEKDKDPAFR 600
601 KQKRNFLQRL FDNWDSAHIG ELTLNDVVTG LDKLVTVDLL QAINYFFSLY DTDGDGELHR 660
661 EEVLQLSEGL LLLTEPWKSG RYVDLLTKKR IEDDIAENII KESGGEIATM NQIELPTGVT 720
721 IDEEKYKVEQ AERYLKAASN FLQRSFEYAK AVDLAEEVNL IDLSDDEGEE KRTVKQKQLE 780
781 SIKANAALDP THPKVIDLPT FRMIILADET YELFFSNTLR SSVHVDEHVN IDNKNKVLRS 840
841 MFDGILADGK RVAEQVRRRV DSVATRSSIA SVESTPTAAA SSITTKEEKY DDLDDFTSEH 900
901 QPENEELLQS SWFEIDDANE TSTKAIQERS FEPLSANSSE EKSNLIEFEA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
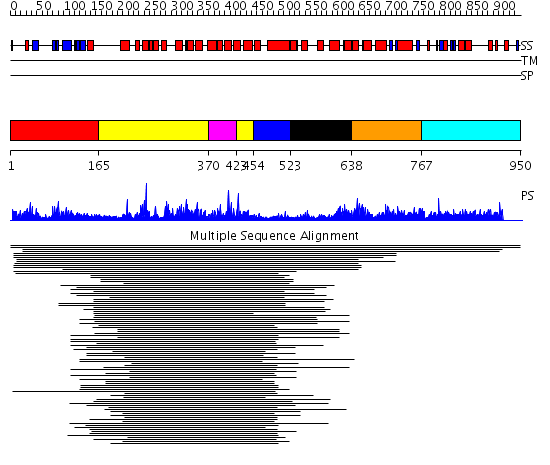
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..164] | 21.420216 | GRAM domain No confident structure predictions are available. | |
| 2 | View Details | [165..369] [423..453] |
43.154902 | Ypt/Rab-GAP domain of gyp1p | |
| 3 | View Details | [370..422] | 43.154902 | Ypt/Rab-GAP domain of gyp1p | |
| 4 | View Details | [454..522] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [523..637] | 57.66807 | Recoverin | |
| 6 | View Details | [638..766] | 57.66807 | Recoverin | |
| 7 | View Details | [767..950] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)