













| General Information: |
|
| Name(s) found: |
gi|190691485
[NCBI NR]
gi|190690113 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 664 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METPSQRRAT RSGAQASSTP LSPTRITRLQ EKEDLQELND RLAVYIDRVR SLETENAGLR 60
61 LRITESEEVV SREVSGIKAA YEAELGDARK TLDSVAKERA RLQLELSKVR EEFKELKARN 120
121 TKKEGDLIAA QARLKDLEAL LNSKEAALST ALSEKRTLEG ELHDLRGQVA KLEAALGEAK 180
181 KQLQDEMLRR VDAENRLQTM KEELDFQKNI YSEELRETKR RHETRLVEID NGKQREFESR 240
241 LADALQELRA QHEDQVEQYK KELEKTYSAK LDNARQSAER NSNLVGAAHE ELQQSRIRID 300
301 SLSAQLSQLQ KQLAAKEAKL RDLEDSLARE RDTSRRLLAE KEREMAEMRA RMQQQLDEYQ 360
361 ELLDIKLALD MEIHAYRKLL EGEEERLRLS PSPTSQRSRG RASSHSSQTQ GGGSVTKKRK 420
421 LESTESRSSF SQHARTSGRV AVEEVDEEGK FVRLRNKSNE DQSMGNWQIK RQNGDDPLLT 480
481 YRFPPKFTLK AGQVVTIWAA GAGATHSPPT DLVWKAQNTW GCGNSLRTAL INSTGEEVAM 540
541 RKLVRSVTVV EDDEDEDGDD LLHHHHGSHC SSSGDPAEYN LRSRTVLCGT CGQPADKASA 600
601 SGSGAQVGGP ISSGSSASSV TVTRSYRSVG GSGGGSFGDN LVTRSYLLGN SSPRTQSPQN 660
661 CSIM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
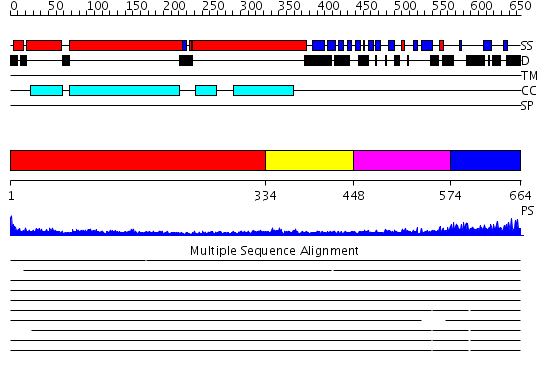
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..333] | 15.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [334..447] | 5.30103 | No description for 2dfsA was found. | |
| 3 | View Details | [448..573] | 15.045757 | Lamin A/C globular tail domain | |
| 4 | View Details | [574..664] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
|||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)